
Immune infiltration, glioma stratification, and therapeutic implications
Glioma: classification, prognosis
By definition, gliomas are brain tumors emanating from glial precursor cells. Gliomas include glioblastoma (GBM), astrocytoma, oligodendroglioma, and ependymoma. The World Health Organization (WHO) traditionally classifies brain tumors according to their pathological features, with GBM having the highest grade (IV), while lower grades (mostly II, III) are typically assigned to various types of astrocytic, oligodendroglial, and ependymal tumors (1), which collectively are referred to as lower grade gliomas (LGG). In the latest NCI SEER (Surveillance, Epidemiology, and End Results) report and projection, the estimated 2016 incidence and deaths for brain and other CNS tumors is 23,770 and 16,050, respectively (2). According to CBTRUS (Central Brain Tumor Registry of the United States), glioma represents about 27% of all, and 80% of malignant brain and CNS tumors (3). The majority (55.1%) of gliomas are classified as GBMs. For GBMs, the current standard of care consists of surgical resection, radiotherapy with chemotherapy (temozolomide or TMZ). The median survival rate, which ranges from 14.5 to 16.6 months [see (4,5) for review], remains dismally poor despite decades of research and discoveries, leading up to the current era of genomics, targeted therapeutics, and immunotherapeutics. LGGs on the other hand have more favorable clinical outcome, especially cases possessing IDH mutation and co-deletion of 1p and 19q chromosomal arms.
The molecular classifications of gliomas
Data generated through various genome-wide molecular profiling tools (e.g., mRNA levels, gene copy number, CpG methylation, mutation profile) led to the molecular classifications of tumors. Analysis of TCGA GBM expression data led to the identification of four molecular subtypes (Proneural, Neural, Classical and Mesenchymal) (6). More recent analyses of TCGA LGG genomic have identified three subtypes with molecular signatures aligned with a tumor’s IDH, 1p/19q, and TP53 status (7).
The immune infiltrate signatures of gliomas
Doucette and colleagues conducted further analysis on the TCGA GBM expression dataset and have concluded that the four molecular subtypes also exhibit differing immune signatures (8). In particular, the authors concluded that the mesenchymal subtype is characterized by a pro-inflammatory signature, and immunosuppression. Cheng and colleagues (9), using a different bioinformatic approach recently described the association of immune infiltrates to higher risk (i.e., poorer clinical outcome) in gliomas. Through Univariate Cox Regression Analysis, they were able to identify eight genes whose expression are found to correlate best with prognosis in glioma. This was accomplished using expression data (for 322 immune-related genes) from 297 tumor samples from Chinese Glioma Genome Atlas (CGGA), as well as the TCGA GBM expression dataset for validation. The elevated expression of the genes CCL18 (C-C motif chemokine ligand 18), MMP9 (matrix metallopeptidase 9), FCGR2B (Fc fragment of IgG receptor IIb), IL6 (interleukin 6), and IL10 (interleukin 10), are associated with poor prognosis (hazard ratio >1), while those of FOXO3 (forkhead box O3), AIMP1 (aminoacyl tRNA synthetase complex interacting multifunctional protein 1), and ZBTB16 (zinc finger and BTB domain containing 16) associate with good prognosis (hazard ratio <1). The regression formula for Risk Score (RS) is: −0.6718 (FOXO3) + 0.1658 (IL6) + 0.2584 (IL10) − 0.1811 (ZBTB16) + 0.1165 (CCL18) − 0.4046 (AIMP1) + 0.1543 (FCGR2B) + 0.1223 (MMP9), where each gene symbol represents expression level. Further analyses would show that higher RS is related to the mesenchymal subtype, lower overall survival, likely wild type IDH1, and lower degree of MGMT promoter methylation among GBMs. The authors then divided each of the glioma cohort across RS median (below the median is low risk or LR, while above the median is high risk or HR), and by employing GSEA (Gene Set Enrichment Analysis) (10), they demonstrated that the Gene Ontology (GO) Bioprocess gene sets M13664 (Immune System Process; with 334 member genes) and M1987 (Immune Response; with 237 member genes) exhibit a high Normalized Enrichment Score (NES).
A closer look at the immune-related processes associated with high risk gliomas
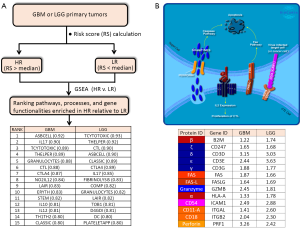
In order to obtain an expanded view of the types of immune processes which are upregulated as Risk Score increases, we ran GSEA (http://software.broadinstitute.org/gsea/index.jsp) on both TCGA GBM (154 primary tumors) and LGG (516 primary tumors) RNASeq-generated expression datasets (downloaded from UCSC Cancer Genomics Browser website: https://genome-cancer.ucsc.edu), against the Biocarta gene sets (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways) (See Figure 1A). Comparisons were made between HR and LR primary tumors. Consistent with the findings by Cheng et al., there is indeed a high degree of immune-related molecular signatures in HR relative to LR cases, for either cohort. These pathways include: ASBCELL (Antigen Dependent B Cell Activation), IL17 (IL 17 Signaling Pathway), TCYTOTOXIC (T Cytotoxic Cell Surface Molecules), THELPER (T Helper Cell Surface Molecules), GRANULOCYTES (Adhesion and Diapedesis of Granulocytes), CTL (CTL mediated immune response against target cells), CTLA4 (The Co-Stimulatory Signal During T-cell Activation), LAIR (Cells and Molecules involved in local acute inflammatory response), IL10 (IL-10 Anti-inflammatory Signaling Pathway), DC (Dendritic cells in regulating TH1 and TH2 Development), and CLASSIC (Classical Complement Pathway). Exemplified in Figure 1B is the CTL pathway, with ES of 0.88 and 0.90 for GBM and LGG, respectively. In this model, the cancer-specific antigen can be presented by the cancer cell through the MHC-1 receptor (indicated in the diagram as red-colored complex of HLA-A (α) and the beta-2-microglobulin (β) chains), to the cytotoxic T cell receptor (represented in the diagram as dark blue-colored complex of highly variable α and β chains, and the ζ, δ, ε, and γ proteins (coded by genes CD247, CD3D, CD3E, and CD3G, respectively). This connection between cytotoxic T cell (CTL) and the tumor cell then leads to eventual apoptosis of the tumor cell through Fas-Fas ligand interactions, and perforin-mediated granzyme activation of the caspase cascade. This interaction also triggers the proliferation of the cytotoxic T cell. The high ES values for this particular pathway can be explained by the elevated transcription (HR relative to LR) of all the genes involved in the process (Figure 1B). As shown in the inset table, genes presumed to be expressed in CTL, such as the T cell receptor genes (CD247, CD3D, CD3E, CD3G), PRF1, GZMB, FASLG, ITGAL, and ITGB2, as well as genes presumably expressed in the antigen-presenting cell, such as ICAM1, HLA-A, B2M, and FAS, are all over-expressed in HR relative to LR cases. The enrichment of the gene set THELPER (T Helper Cell Surface Molecules) is an indication of increased population of T helper cells infiltrating the HR tumors. The primary genetic markers of this particular gene set are the genes coding for the glycoproteins CD54 (ICAM1), CD4, and CD2, as well as T cell receptor component proteins, all of which are upregulated in HR relative to LR tumors. Also enriched in HR gliomas is the gene set for CTLA4 signaling (The Co-Stimulatory Signal During T-cell Activation), a pathway directly relevant to immunotherapy, given that CTLA4 is a primary target for checkpoint inhibition. The enrichment of CTLA4 pathway is driven by the upregulation (HR vs. LR) of genes coding for T cell surface molecules (CTLA4, CD28, ICOS, and the T cell receptor genes), intracellular tyrosine kinases crucial to T cell proliferation (LCK, ITK), as well as proteins on the surface or antigen-presenting cells (CD86, CD80, HLA-DRB1). The enrichment of inflammatory and neutrophil-recruiting IL17 Signaling Pathway is supported by the elevated expression of the cytokines IL6, IL8, and CSF3. The presence of granulocytes around HR tumors is supported by the elevated level of granulocyte surface molecules (ITGAM/CD11B, ITGB2/CD18, SELL/L-selectin), and the stimulating cytokines (IL8, CSF3, IL1A, TNF). Also likely to be more prominent around HR glioma cells are dendritic cells, needed for the stimulation of lymphocytes. The gene set DC (Dendritic cells in regulating TH1 and TH2 Development) acquired high ES due to the upregulation of IL10, CSF2, CD2, CD7, CD33, and CD40. The upregulation of complement components C7, C1S, C1QA, C2 explains why the gene set Classic (Classical Complement Pathway) is enhanced in HR relative eto LR glioma. The activation of B cells around HR gliomas is also likely as indicated by the enrichment of the gene set ASBCELL, with most of its component genes (IL10, CD28, CD80, FASLG, FAS, CD40, HLA-DRB-1, CD40LG) over-expressed in HR compared to LR glioma.
Implications in immunotherapy
What we illustrated here is that in glioma (GBM or LGG), molecular pathways pertaining to immune infiltration (e.g., CD4+ lymphocytes, Dendritic cells, Granulocytes, inflammation) may tend to be more pronounced among tumors with higher RS (9) (and lower overall survival). In GBMs, higher RS is also associated with the mesenchymal molecular subtype. In an earlier clinical study involving 23 GBM patients, Prins and colleagues have shown that tumor samples exhibiting mesenchymal molecular signature, indeed have higher population of both CD3+ and CD8+ lymphocytes (11), though others are of the opinion that increased tumor infiltration of CD3+ and CD8+ cells correlates with prolonged patient survival (12), and high level of CD4+ tumor infiltrating lymphocytes combined with low CD8+ tumor infiltrating lymphocytes is reported to be associated with unfavorable prognosis (13). Such studies also led to another important observation, that mesenchymal (thus more highly infiltrated) samples were more responsive to dendritic cell vaccination. It is quite plausible that gliomas with higher degree of immune infiltration (thus higher RS) may also respond more positively to checkpoint inhibitors such as the anti-CTLA4 ipilimumab and the anti-PD1 nivolumab and pembrolizumab. Ongoing clinical trials of these drugs against glioma include: NCT02311920 (phase I; ipilimumab and/or nivolumab in combination with temozolomide against newly diagnosed GBM or gliosarcoma), NCT02337491 (phase II; pembrolizumab with or without bevacizumab against recurrent GBM), NCT02017717 (phase III; effectiveness and safety of nivolumab compared to bevacizumab and of nivolumab with or without ipilimumab in GBM) [see (14), or clinicaltrials.gov]. It is worth noting that the fold-change (HR vs. LR) for CTLA4 is 1.6 and 1.7 for GBM and LGG, respectively, while the fold-change (HR vs. LR) for PD1 is 1.6 and 2.4 for GBM and LGG, respectively.
Acknowledgments
Funding: Research support provided in part by National Institutes of Health, National Cancer Institute 1R01 CA134721 (PB Fisher), NIH/NCI Massey Cancer Support Grant 5P30 CA016059 (PB Fisher, D Sarkar, XY Wang), the National Foundation for Cancer Research (NFCR) (PB Fisher) and Department of Defense Synergy Grant W81XWH-14-1-0409 (PB Fisher, XY Wang). PB Fisher holds the Thelma Newmeyer Corman Chair in Cancer Research.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Ning Huang (Department of Neurosurgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.69). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol 2015;17:iv1-iv62. [Crossref] [PubMed]
- Wen PY, Reardon DA. Neuro-oncology in 2015: Progress in glioma diagnosis, classification and treatment. Nat Rev Neurol 2016;12:69-70. [Crossref] [PubMed]
- Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98-110. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372:2481-98. [Crossref] [PubMed]
- Doucette T, Rao G, Rao A, et al. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res 2013;1:112-22. [Crossref] [PubMed]
- Cheng W, Ren X, Zhang C, et al. Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology 2016;86:2226-34. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Prins RM, Soto H, Konkankit V, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res 2011;17:1603-15. [Crossref] [PubMed]
- Kmiecik J, Poli A, Brons NH, et al. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J Neuroimmunol 2013;264:71-83. [Crossref] [PubMed]
- Han S, Zhang C, Li Q, et al. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br J Cancer 2014;110:2560-8. [Crossref] [PubMed]
- Binder DC, Davis AA, Wainwright DA. Immunotherapy for cancer in the central nervous system: Current and future directions. Oncoimmunology 2015;5:e1082027 [Crossref] [PubMed]


