
The interaction between TopBP1 and MCM complex is essential for TopBP1 foci formation in colon cancer
Introduction
Topoisomerase IIβ-binding protein 1 (TopBP1) is a checkpoint protein, which involves in response to DNA damage and associates with the chromosome replication (1,2). TopBP1 localizes to the mitotic centrosome and mediates progression of mitosis (3). TopBP1 is a key player in homologous recombination and plays a role through interaction with other proteins that is related to DNA replication and cancer progression, such as RAD51 (4) and breast cancer susceptibility gene 1 (BRCA1) (5).
BRCA1 is important for maintaining genome stability and suppressing tumor cell function via controlling cell cycle checkpoint and repairing DNA damage (6). BRCA1 C-terminal (BRCT) domains are found in a number of proteins that are involved in cell cycle checkpoint and are essential for BRCA1 DNA repair and its tumor suppressor activity (7). TopBP1 contains eight or nine BRCT domains (3,8). The stimulation of TopBP1 on ataxia-telangiectasia mutated—and Rad3-related (ATR) activity was activated by BRCT7+8 domains of TopBP1 (8,9), partially via ATR-interacting protein (ATRIP)-TopBP1 interaction and activation of ATR-ATRIP (9,10). ATR and ATRIP are mutually dependent on each other for checkpoint signaling (11). TopBP1 activates ATR via ATRIP (9), and ATR-ATRIP binding proteins are vital for the cellular response to DNA damage and replication (11). Aberrant expression of TopBP1 could be detected in various cancers, such as breast cancer (12), lung cancer (13) and ovarian cancer (14).
ATR is mainly localized on chromatin and this chromatin association is essential for the initiation of cell cycle checkpoints (3,15), as well as minichromosome maintenance protein (MCM) complex (16). MCM complex are direct targets of the ATR checkpoint kinase (17), and also required for the elongation of chromosomal DNA replication (18). Moreover, the analysis of MCMs was regarded as a novel method for the colorectal cancer detection (19), and MCMs were considered to be tumor markers for many human tumors, including glioma (20), ovarian cancer (21), breast cancer (22), and Dukes C colorectal cancer (23). Although MCM complex is a core element of DNA replication (15), and MCM complex was indicated as a crucial player in replication checkpoint (24,25), there was little information showing the association of MCMs and other DNA damage response proteins such as TopBP1, ATR and BRCA1.
Given MCMs are tumor markers for colon cancer (19,23), we investigated the interaction between TopBP1 and MCMs in HCT116 colon cancer cell line. HCT116 cells stably expressing Flag-TopBP1 were constructed and fractionated. After chromatin fraction, immunoprecipitation (IP), Western blotting, and in vitro Glutathione S-transferase (GST) pull-down assay, and mass spectrometry (MS) were performed to detect the potential interactions between TopBP1 and MCMs. Moreover, the expression of MCM2 and MCM6 was knocked down by specific short hairpin RNA (shRNA), and then the chromatin fraction and foci formation of TopBP1 were assessed under the condition of DNA damage. This study might provide a new insight into the potential interaction between TopBP1 and MCMs.
Methods
Cells, culture conditions and transfection
HCT116 cells (ATCC, Manassas, VA, USA) were incubated in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco; Thermo Fisher Scientific, Inc., USA) with 10% fetal bovine serum (FBS, Gibco), 100 U/mL of streptomycin and penicillin (Gibco) at 37 °C with 5% CO2. HCT116 cells were transfected by using Lipofectamine 2000 (Invitrogen, CA, USA).
Plasmids, antibodies and reagents
Plasmids of Myc- and green fluorescent protein (GFP)-tagged vectors expressing TopBP1 wide type (WT) and mutants were constructed as previously described by Han et al. (15). MCMs (MCM2, 3, 5, and 6) complementary DNAs (cDNAs) were generated by standard RT-PCR and cloned into pcDNA3.1-HA or Flag mammalian expression vectors (HA-MCMs or Flag-MCMs). Antibodies including anti-TopBP1 (Abcam, Cambridge, USA), anti-Flag (Sigma-Aldrich Co, St. Louis, MO, USA), anti-HA (Roche Diagnostics, Indianapolis, IN), anti-GST (Nacalai Tesque, Japan), and anti-MCMs (Sigma-Aldrich Co, St. Louis, MO, USA) were used for Western blotting and IP. Anti-α-tubulin (Sigma-Aldrich Co, St. Louis, MO, USA) was used as a loading control for Western blotting. Chemically synthesized shRNAs against MCMs were provided by Invitrogen (Carlsbad, CA, USA).
Chromatin fraction
Cell fractionation and chromatin fraction were performed as previously described (15). Briefly, cells were harvested, lysed and centrifuged at 1,000 rpm, 4 °C for 5 min. Then the chromatin-enriched pellet was resuspended with corresponding solutions (15) on ice and centrifuged at 4 °C for three times. The final pellets of chromatin-enriched fraction were resuspended. The supernatant after the second centrifugation was non-chromatin fraction (cytoplasmic fraction). Cell fractions were neutralized for further IP and immunoblot analysis.
IP and Western blotting
For IP determination, cells were lysed, sonicated, and centrifuged as described by Han et al. (15) and elsewhere (26). The supernatants were then treated with anti-TopBP1 antibody overnight. Then protein A/G agarose beads (Amersham Pharmacia Biotech, Rainham, USA) were added and additionally incubated for 2–4 h. The beads were washed three times with lysis buffer and Western blotting was subsequently carried out. In brief, cell lysates (Inputs) or immunoprecipitated proteins were separated by sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA), which were then blocked in blocking solution. The membranes were incubated with primary and secondary antibodies. Next, the immunoreactive protein bands were visualized using enhanced chemiluminescence (ECL+; Amersham Biosciences, Piscat-away, NJ, USA).
GST pull-down assay
In vitro binding assays, GST pull-down, were performed with purified TopBP1 and GST-α proteins bounding to glutathione-agarose beads (27). GST fusion proteins of TopBP1 and GST-α were prepared and were subjected to SDS-PAGE. Cell lysates were used as inputs.
MS assay
HCT116 cells stably expressing Flag-TopBP1 were fractionated into cytoplasmic and nuclear compartments. Identification of TopBP1-interacting protein was completely performed as described by Han et al. (15). Also, SEQUEST search results were assembled and filtered using the DTASelect (version 2.0) algorithm (18), requiring peptides to be at least half-tryptic and a minimum of two peptides per protein identification. The protein identification false positive rate was kept below 5%.
Immunofluorescence microscopy
Before immunofluorescence microscopy, transfected cells were treated with bleomycin (BLM, 2 µM, 1 h; Sigma-Aldrich Co, St. Louis, MO, USA) to induce chromosomal aberration or breaks (28). For microscopy, cells were fixed with paraformaldehyde (PFA) and permeabilized sequentially with ethanol on ice (29). The slides were blocked and incubated with primary antibodies anti-TopBP1. Thereafter, the cells were washed and incubated with biotin-conjugated secondary antibodies Cell Signaling Technology, Beverly, MA) for 2 h at room temperature. DNA was stained with 4’,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). The slips were observed on a fluorescence microscope (Olympus Power BX51, Olympus, Co., Tokyo, Japan) and the data were analyzed by NIH ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All experiments were performed in triplicate. The data for TopBP1 positive cells of immunofluorescence microscopy are expressed as means ± standard deviations (SD). P<0.05 was considered statistically significant. Comparisons between groups were analyzed by using Statistic Package for Social Science version 18.0 (SPSS; SPSS Inc., Chicago, IL, USA).
Results
TopBP1 interacts with the MCMs
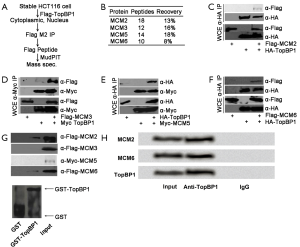
To determine the functional roles of TopBP1 in cells, we intended to explore the specific TopBP1-interacting proteins. In this study, we identified the specific TopBP1-interacting proteins in the nucleus of HCT116 cells stably expressing FLAG-TopBP1 (Figure 1A). Furthermore, we identified four MCMs, MCM2, 3, 5, and 6, interacted with TopBP1 (Figure 1B). IP and Western blotting were performed to determine the interaction between TopBP1 and MCM proteins. The results demonstrated that TopBP1 interacted with these four MCMs (Figure 1C-F). Moreover, the in vitro GST pull-down assay were performed with purified TopBP1 and GST-α proteins bounding to glutathione-agarose beads to identify the results. As expected, we detected there were interactions between TopBP1 and MCM2 and MCM6. However, there were no interactions between TopBP1 and MCM3 and MCM5 (Figure 1G). To further verify the interaction between endogenous TopBP1 and MCM proteins, protein lysates were prepared from HCT116 cells (without transfection), followed by IP by using a Co-Immunoprecipitation Kit (Pierce, Carlsbad, CA, USA) according to the manufacturer’s instructions. IP lysates were incubated with anti-TopBP1. IgG was used as a negative control, and while input (without antibody) was considered as a positive control. Thereafter, WB was performed to detect the IP products. The detection results showed that the protein levels of MCM2, MCM6, and TopBP1 were all increased by incubation with anti-TopBP1 compared to the negative control, indicating that there was an interaction between endogenous TopBP1 and MCM proteins (Figure 1H).
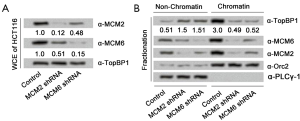
Downregulation of MCMs reduces TopBP1 in chromatin fraction
Evidence from other groups showed that human TopBP1 was localized to the mitotic centrosome and mediated progression of mitosis (3). TopBP1 is a checkpoint protein and had been reported to be involved in response to DNA damage, and also to be associated with the chromosome replication (1,2). Since research had shown that TopBP1 interacted with MCM2 and MCM6, and TopBP1 was involved in DNA damage response, we supposed that MCM2 and MCM6 might play an important role in TopBP1 chromatin localization and DNA damage. To confirm the supposition, we downregulated the expression of MCMs via shRNA and successfully suppressed the expression of MCM2 and MCM6 ranging from 50% to 80% (Figure 2A). Then we determined the expression of TopBP1 in chromatin compartments, compared with non-chromatin fraction compartments. As shown in the results, there was a significant reduction of TopBP1 level in chromatin compartment compared to the non-chromatin fraction compartments, along with an increase of TopBP1 level in non-chromatin compartment (Figure 2B).
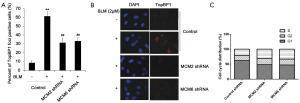
Downregulation of MCMs reduces TopBP1 foci formation
Previous studies have shown that BLM could induce the chromosomal aberration or breaks (28) as well as the foci formation of DNA damage response genes (30). In addition, TopBP1 is involved in response to DNA damage (1,2), BLM-TopBP1 could maintain genome stability (31), and TopBP1 interacted with MCM2 and MCM6. We therefore speculated whether MCMs were essential for TopBP1 foci formation. To verify this question, we first down-regulated the expression of MCM2 and MCM6 in HCT116 cells using shRNA, then treated cells with 2 µM BLM for 1 h, and observed the expression of TopBP1 by immunofluorescence microscopy. From the data of immunofluorescence microscopy, we declared that BLM administration induced the chromosomal aberration and TopBP1 foci formation (Figure 3A,B). After silencing of MCM2 and MCM6, the percentages of TopBP1 positive cells were significantly reduced (P<0.01). This results demonstrated that MCMs were essential for TopBP1 foci formation, and downregulation of MCMs could reduce BLM induced TopBP1 foci formation. Further, we analyzed the cell cycle distribution after silencing of MCM2 and MCM6. As shown in Figure 3C, the results showed that the percentages of cells in S phase were significantly increased after knockdown of MCM2 and MCM6, indicating that the TopBP1-foci formation was largely restricted to S phases.
Discussion
This study provided evidence for a novel interaction between TopBP1 and MCMs, especially MCM2 and MCM6. MS, IP, Western blotting, and GST pull-down assay were performed and confirmed the interactions between TopBP1 and MCMs, including MCM2, 3, 5, and MCM6. Moreover, we demonstrated that MCM2 and MCM6 were essential for TopBP1 chromatin localization and TopBP1 foci formation in HCT116 cells. Downregulation of MCMs reduced TopBP1 foci formation and chromatin fraction.
TopBP1 is a checkpoint protein which mediates progression of mitosis (3). TopBP1 has been determined to be localized to the mitotic centrosome and involved in response to DNA damage, and associated with the chromosome replication (1,2). Moreover, MCM complex is a core element for DNA replication (15). MCM complex was indicated as a crucial player in replication checkpoint (24,25). However, the interaction between TopBP1 and MCMs had rarely been reported. In this study, we firstly identified the interaction between TopBP1 and MCMs, including MCM2, MCM3, MCM5 and MCM6. The results suggested that TopBP1-MCM2 or -MCM6 bounding proteins might play great roles in DNA damage or chromosome replication.
At the beginning of mitosis, obvious accumulation of TopBP1 on chromatin is observed, which gradually dissociates during mitosis progression (32). TopBP1 contains eight or nine BRCT domains encoding damage response proteins which are found in proteins involved in cell cycle checkpoint (3,8). The stimulation of TopBP1 on ATR activity is activated by BRCT 7+8 domains (8-10), partially via ATRIP-TopBP1 interaction (9). ATR is mainly localized on chromatin and chromatin association, which is essential for the initiation of cell cycle checkpoints and TopBP1 accumulation on chromatin (3,15). In this study, we observed that TopBP1 interacted with the MCM complex and this interaction was decreased by DNA damage, which led us to assume whether the MCM complex is responsible for the recruitment of TopBP1 to chromatin. To confirm the assumption, we downregulated expression of MCM2 and MCM6 in HCT116 cells using MCMs shRNA for 72 h, and then we determined the expression of TopBP1 in chromatin compartments. A clear reduction of TopBP1 level in chromatin compartment was detected, as well as an increase of TopBP1 level in non-chromatin compartment. This proved that MCM2 and MCM6 were essential for TopBP1 on chromatin localization, and downregulation of MCMs reduced TopBP1 chromatin fraction. Moreover, our results might provide basic information on the positive association between MCMs with ATR and cell cycle checkpoint (24). However, it should be noted that following depletion of MCM2 and 6 for 72 h, the cells are cell cycle arrested. This might be one of the reasons to explain the shift of TopBP1 from chromatin to soluble fraction.
Chromosomal break is a prerequisite and chromosome or DNA replication (33) and cell cycle, and one source of nonreciprocal translocations and telomere capture (34). BLM could induce the chromosomal aberration or breaks (28) as well as the foci formation of DNA damage response genes, such as histone variant H2AX (‘γH2AX’), ATM and p53-binding protein 1 (53BP1) (30,35). As reported, γH2AX plays an important role in sensing and repairing DNA damage. Phosphorylation of H2AX histone is linearly related to an early event of DNA double-strand break (DSB) (30). Chromosomal breaks during mitosis might trigger γH2AX mediated apoptosis (36), resulting in nonreciprocal chromosomal translocations after recombination (34) and inducing genetic variation, genetic information loss or diseases progression (37,38). In this study, the fact that TopBP1 protein was upregulated in BLM treated HCT116 cells demonstrating TopBP1 was involved in response to DNA damage and chromosomal break (1,2). Moreover, we suggested that BLM-induced TopBP1 foci formation could be inhibited by MCMs reduction. There was reduction of TopBP1 protein in chromatin fraction compartments of MCM2 and MCM6 shRNA cells and a concomitant upregulation in non-chromatin fraction compartments of MCM2 and MCM6 shRNA cells. Additionally, reduction of BLM-induced TopBP1 foci positive cells number was detected in MCM2 and MCM6 shRNA cells. These results suggested that MCM2 and MCM6 were essential for progression of mitosis by interacting with or regulating TopBP1. However, we should also note that the cells might be arrested at a particular phase of the cell cycle following MCM depletion, which could equally explain the reduction of TopBP1 foci formation. To confirm the results, we analyzed the cell cycle distribution after silencing of MCM2 and MCM6. The results demonstrated that knockdown of MCM2 and MCM6 significantly increased the percentages of S phase, suggesting that TopBP1-foci formation was largely restricted to S phases. Taken together, the results in this study confirmed that TopBP1 might play a crucial role in the cell cycle and DNA replication, and that suppression of MCMs reduces the chromatin fraction and foci formation of TopBP1 upon DNA damage in HCT116 cells. The potential interactions might provide a new insight into improve the outcome of chemotherapy in colon cancer. However, further studies should be performed to confirm the relationship between TopBP1 and MCMs.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.51). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herold S, Hock A, Herkert B, et al. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J 2008;27:2851-61. [Crossref] [PubMed]
- Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 2005;4:1227-39. [Crossref] [PubMed]
- Bang SW, Ko MJ, Kang S, et al. Human TopBP1 localization to the mitotic centrosome mediates mitotic progression. Exp Cell Res 2011;317:994-1004. [Crossref] [PubMed]
- Moudry P, Watanabe K, Wolanin KM, et al. TOPBP1 regulates RAD51 phosphorylation and chromatin loading and determines PARP inhibitor sensitivity. J Cell Biol 2016;212:281-8. [Crossref] [PubMed]
- Greenberg RA, Sobhian B, Pathania S, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev 2006;20:34-46. [Crossref] [PubMed]
- Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 2010;11:138-48. [Crossref] [PubMed]
- Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol 2001;8:838-42. [Crossref] [PubMed]
- Wardlaw CP, Carr AM, Oliver AW. TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair (Amst) 2014;22:165-74. [Crossref] [PubMed]
- Mordes DA, Glick GG, Zhao R, et al. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 2008;22:1478-89. [Crossref] [PubMed]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003;300:1542-8. [Crossref] [PubMed]
- Going JJ, Nixon C, Dornan ES, et al. Aberrant expression of TopBP1 in breast cancer. Histopathology 2007;50:418-24. [Crossref] [PubMed]
- Kumagai A, Lee J, Yoo HY, et al. TopBP1 activates the ATR-ATRIP complex. Cell 2006;124:943-55. [Crossref] [PubMed]
- Choi SH, Yang H, Lee SH, et al. TopBP1 and Claspin contribute to the radioresistance of lung cancer brain metastases. Mol Cancer 2014;13:211. [Crossref] [PubMed]
- Karppinen SM, Erkko H, Reini K, et al. Identification of a common polymorphism in the TopBP1 gene associated with hereditary susceptibility to breast and ovarian cancer. Eur J Cancer 2006;42:2647-52. [Crossref] [PubMed]
- Han X, Aslanian A, Fu K, et al. The interaction between checkpoint kinase 1 (Chk1) and the minichromosome maintenance (MCM) complex is required for DNA damage-induced Chk1 phosphorylation. J Biol Chem 2014;289:24716-23. [Crossref] [PubMed]
- Nedelcheva MN, Roguev A, Dolapchiev LB, et al. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J Mol Biol 2005;347:509-21. [Crossref] [PubMed]
- Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci U S A 2004;101:10078-83. [Crossref] [PubMed]
- Lee JK, Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc Natl Acad Sci U S A 2001;98:54-9. [Crossref] [PubMed]
- Davies RJ, Freeman A, Morris LS, et al. Analysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stool. Lancet 2002;359:1917-9. [Crossref] [PubMed]
- Hua C, Zhao G, Li Y, et al. Minichromosome Maintenance (MCM) Family as potential diagnostic and prognostic tumor markers for human gliomas. BMC Cancer 2014;14:526. [Crossref] [PubMed]
- Kobierzycki C, Pula B, Skiba M, et al. Comparison of minichromosome maintenance proteins (MCM-3, MCM-7) and metallothioneins (MT-I/II, MT-III) expression in relation to clinicopathological data in ovarian cancer. Anticancer Res 2013;33:5375-83. [PubMed]
- Kwok HF, Zhang SD, McCrudden CM, et al. Prognostic significance of minichromosome maintenance proteins in breast cancer. Am J Cancer Res 2014;5:52-71. [PubMed]
- Ishibashi Y, Kinugasa T, Akagi Y, et al. Minichromosome maintenance protein 7 is a risk factor for recurrence in patients with Dukes C colorectal cancer. Anticancer Res 2014;34:4569-75. [PubMed]
- Tsai FL, Vijayraghavan S, Prinz J, et al. Mcm2-7 Is an Active Player in the DNA Replication Checkpoint Signaling Cascade via Proposed Modulation of Its DNA Gate. Mol Cell Biol 2015;35:2131-43. [Crossref] [PubMed]
- Han X, Mayca Pozo F, Wisotsky JN, et al. Phosphorylation of Minichromosome Maintenance 3 (MCM3) by Checkpoint Kinase 1 (Chk1) Negatively Regulates DNA Replication and Checkpoint Activation. J Biol Chem 2015;290:12370-8. [Crossref] [PubMed]
- Donaldson MM, Mackintosh LJ, Bodily JM, et al. An interaction between human papillomavirus 16 E2 and TopBP1 is required for optimum viral DNA replication and episomal genome establishment. J Virol 2012;86:12806-15. [Crossref] [PubMed]
- Murata K, Wu J, Brautigan DL. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc Natl Acad Sci U S A 1997;94:10624-9. [Crossref] [PubMed]
- Mira A, Gimenez EM, Bolzán AD, et al. Effect of thiol compounds on bleomycin-induced DNA and chromosome damage in human cells. Arch Environ Occup Health 2013;68:107-16. [Crossref] [PubMed]
- Nakanishi A, Han X, Saito H, et al. Interference with BRCA2, which localizes to the centrosome during S and early M phase, leads to abnormal nuclear division. Biochem Biophys Res Commun 2007;355:34-40. [Crossref] [PubMed]
- Sánchez-Flores M, Pásaro E, Bonassi S, et al. γH2AX assay as DNA damage biomarker for human population studies: defining experimental conditions. Toxicol Sci 2015;144:406-13. [Crossref] [PubMed]
- Blackford AN, Nieminuszczy J, Schwab RA, et al. TopBP1 interacts with BLM to maintain genome stability but is dispensable for preventing BLM degradation. Mol Cell 2015;57:1133-41. [Crossref] [PubMed]
- Pedersen RT. The Functional Role of TopBP1 in DNA Maintenance at Mitosis. Copenhagen: University of Copenhagen, 2015.
- Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol 2013;5:a010397 [Crossref] [PubMed]
- Bosco G, Haber JE. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 1998;150:1037-47. [PubMed]
- Honnenahally K, Shi C, Chen X, et al. γH2AX: A molecular marker of DNA damage response in smoking-induced pancreatic ductal adenocarcinoma. Cancer Res 2015;75:A33. [Crossref]
- Imreh G, Norberg HV, Imreh S, et al. Chromosomal breaks during mitotic catastrophe trigger γH2AX-ATM-p53-mediated apoptosis. J Cell Sci 2011;124:2951-63. [Crossref] [PubMed]
- Di Virgilio M, Callen E, Yamane A, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 2013;339:711-5. [Crossref] [PubMed]
- Samans B, Chalhoub B, Snowdon R. Homeologous Non-Reciprocal Translocations (HNRT) Induce Selectable Genetic Variation in Brassica napus. Plant & Animal Genome, San Diego, January 10-14, 2015.


