
miR-448 suppresses E2F3 to inhibit proliferation and induce apoptosis in human colorectal cancer cells
Introduction
Colorectal cancer (CRC) is the one of the most commonly diagnosed malignancies and accounts for one of the major global health problems (1,2). The molecular basis of the tumorigenesis of CRC is still incompletely discovered, limiting the development of new effective targeted therapeutic drugs. It is therefore necessary to study the pathogenesis of CRC at the molecular level.
MicroRNAs (miRs) refer to a subclass of non-coding RNAs, which are about 20 nt in length and negatively affect gene expression at the posttranscriptional level (3). Since its discovery, miRs have caught wide attentions both in the research and clinic filed, as various miRs have been demonstrated to be involved in a large set of biological processes (4,5). Abnormal expression of miRs in cancer cells often has a causal role of the hyperproliferative ability of the tumor cells, and in CRC, various miRs have been identified as oncogenic or tumor suppressive factors (6). It has been reported that upregulation of miR-21 plays an oncogenic role (7); and p53 guided miR-34a expression suppresses tumor progression (8). It is anticipated that new mechanisms of the posttranscriptional regulation by other miRs may also contribute greatly to the pathogenesis of CRC.
In this study, we asked the possible function of the seldom investigated miR-448 in CRC. Pilot studies have shown that this microRNA negatively regulates cell growth in gastric cancer and ovarian cancer (9,10), which implied the possible role of miR-448 in CRC. To this end, we firstly measured whether miR-448 is differentially expressed in CRC tissues, and then we asked what does miR-448 function in regulating tumor cell activity and through what mechanism does miR-448 change the tumor biology of CRC. Our study for the first time revealed the tumor suppressive role of miR-448 and established its direct interaction with E2F3 transcriptional factor in CRC.
Methods
Tumor specimens
Thirty-four paired CRC specimens were collected from surgery. The tissues were diagnosed by experienced pathologist. The tissues were transferred to liquid nitrogen immediately. None of the patients underwent adjuvant therapy such as chemotherapy and radiotherapy before surgical resection. The study was approved by the ethics committee of the affiliated hospital of Qingdao university (approval NO. QYFYEC 2016-012-01) and written informed consent was obtained from all patients.
Cell culture
The human CRC cell lines SW480 and HCT116 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were incubated in a humidified atmosphere at 37 °C. Cells were cultured with RPMI-1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% Fetal Bovine Serum (FBS, Hyclone).
MicroRNAs and plasmid transfection
miR-448 mimics, antimiR-448 and negative controls were purchased from Ribobio Technology (Guangzhou, China). To construct plasmid overexpressing E2F3, the E2F3 coding sequence was cloned from the cDNA by PCR and inserted into the multiple cloning site of the pcDNA3.1 backbone. Cells grown at 80% confluence were subjected to transfection using Lipofectamin 2000 (Invitrogen, Carlsbad, CA, USA) as per manufacturer’s instruction. The transfection concentration was 100 nM for microRNAs and 1 µg/mL for plasmid. Cells were maintained for 12 h after adding the transfection mix, and the media were then refreshed before further experiments.
Cell viability assay
Cell viability was determined by MTT assay. Briefly, cells were plated into a 96-well plate at the concentration of 2×104/mL. After transfection, cells were cultured for 24, 72, and 96 h. MTT reagent (5 mg/mL, 20 µL/well) was added to the culture medium and incubated with the cells for 4 h. Then 200µl DMSO was added to each well and the plates were shaked for 15 min at room temperature. The optical density value at 490 nm was collected using a microplate reader (Biotek, Winooski, VT, USA).
BrdU (5-bromo-2'-deoxyuridine) proliferation assay
Cells were plated on coverslips and transfected as described above. Cells proliferation was determined using a BrdU labeling kit (Roche, Indianapolis, IN, USA). The BrdU incorporation was detected by FITC-BrdU antibody (Roche) and analyzed under a fluorescent microscope. The experiments were performed in triplicate.
Apoptosis assay
Apoptosis rate was determined by transferase-mediated deoxyuridine triphosphate-biotin nick end labeling (TUNEL) assay (Roche, Indianapolis, IN, USA). Cells grown on coverslips were transfected as described for 72 h, followed by fixing with 4% paraformaldehyde (PFA). The cells were permeabilized using 0.4% Triton X100-PBS on ice and incubated with TUNEL reaction mixture at 37 °C for 1 h. The coverslips were counterstained with DAPI. The apoptotic cells were then observed under a fluorescent microscope. The apoptosis rate was counted from 3 independent experiments, and 10 microscopic fields were calculated in each slide.
Real-time PCR
Total RNA from CRC samples or cells was extracted by Trizol Reagent (Invitrogen). The RNA samples were reverse transcribed using M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. The cDNA templates were then subjected to PCR amplification using a SYBR Green master mix on a Biorad CFX-96 thermocycler (Hercules, CA, USA). PCR primer sets for miR-448 and its normalization control U6 were purchase from RiboBio Tech. GAPDH was used for normalizing E2F3 expression. The primers for E2F3 were forward AGAAAGCGGTCATCAGTACCT and reverse TGGACTTCGTAGTGCAGCTCT. The primers for GAPDH were forward GGAGCGAGATCCCTCCAAAAT and reverse GGCTGTTGTCATACTTCTCATGG.
Western blot
Whole cell lysates were collected using SDS lysis buffer (Beyotime, Shanghai, China). The protein was quantified by BCA kit (Beyotime) and about 30 µg protein was loaded onto the SDS-polyacrylamide gel (12%). After electrophoresis, proteins were transferred onto a nitrocellulose membrane. The membranes were blocked with 5% non-fat milk in PBS for 2 h at room temperature and washed with PBS for 3 times. The membranes were incubated with rabbit anti human E2F3(Santa Cruz Biotech, CA, USA), GAPDH (Santa Cruz Biotech) and cleaved-PARP (Cell Signaling Technologies Inc., MA, USA) at 4 °C overnight. Membranes were then washed for 5 times with PBST (PBS + 0.5% Tween 20) and incubated with HRP conjugated horse anti rabbit IgG (Santa Cruz). An ECLplus kit (Beyotime) was used to detect the proteins.
Luciferase assay
Luciferase reporter assay was used to validate the predicted target. Briefly, the 3’UTR containing the putative binding site was cloned into the pmiRGLO construct (Promega, Madison, WI, USA). For mutagenesis, TaKaRa MutanBEST Kit (Dalian, China) was used. The plasmid (wild type 3’UTR or mutant 3’UTR) was transfected into cells along with miR-448 or negative control. The luciferase activity was determined with a Dual luciferase detection system (Promega) according to the instruction 36 h post-transfection.
Statistical analysis
All data were expressed as means ± SD, comparison between two groups was determined by Student’s t-test, and comparison between three groups was determined by one way Analysis of Variance (ANOVA) followed by Tukey’s test. The Spearman correlation analysis was used for analysis of clinical samples. P<0.05 was considered statistically significant.
Results
MiR-448 is down-regulated in CRC
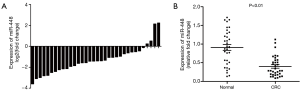
In the first part of the current study, we analyzed whether miR-448 is differentially expressed in CRC. Thirty four paired CRC tissues and adjacent normal tissues were analyzed for miR-448 expression. As shown in Figure 1A, miR-448 expression was down-regulated in most of the CRC tissues (30/34). The mean expression of miR-448 declined by nearly 50%, as compared with that in normal tissues (Figure 1B).
Down-regulation of miR-448 correlates with up-regulation of E2F3
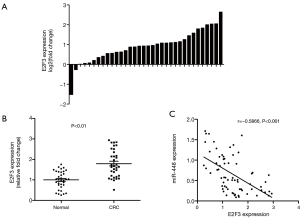
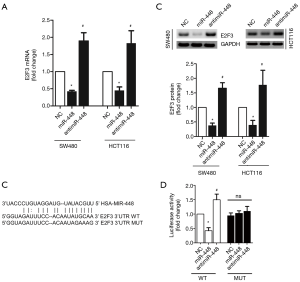
We next assessed the mRNA level of E2F3, an essential gene that controls cell cycle progression. We found that in most of the cases, E2F3 was up-regulated in CRC tissues (31/34) (Figure 2A). And a 2-fold increase of the mean expression of E2F3 was detected (Figure 2B). Importantly, miR-448 expression was inversely correlated with E2F3 expression in the study subjects (Figure 2C), suggesting the potential functional relevance between E2F3 and miR-448.
MiR-448 inhibits cell proliferation and induces apoptosis
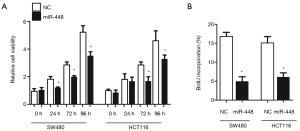
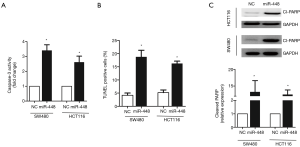
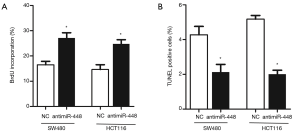
To understand the role of miR-448 in regulating CRC, we employed in vitro experiments to dissect its function. We first transfected miR-448 mimics into SW480 or HCT116 cell line to determine whether miR-448 affect cell proliferation. As shown in Figure 3A, MTT assay revealed that the cell viability was significantly decreased in multiple time points. And less cells were incorporated with BrdU 72 h after transfection (Figure 3B). We next tested whether miR-448 also affects apoptosis. Caspase-3 is a well-known marker of apoptosis and executes apoptosis by cleaving multiple substrates such as PARP. We therefore measured caspase-3 activity, cells overexpressing miR-448 showed a higher level of caspase-3 activity (Figure 4A), and we also confirmed that miR-448 transfected cells exhibited increased TUNEL positive cells (Figure 4B). The cleavage of PARP was also significantly increased (Figure 4C). Conversely, when we transfect CRC cells with the antisense inhibitor of miR-448, we observed a significant increase of cell proliferation (Figure 5A) and a decrease in baseline apoptosis (Figure 5B). These data confirmed that miR-448 inhibits cell proliferation and induces apoptosis in CRC cells.
MiR-448 directly targets E2F3
To understand the molecular mechanism by which miR-448 regulates cell proliferation and apoptosis, we analyzed whether miR-448 and E2F3 were indeed functionally linked. As shown in Figure 6A,B, overexpression of miR-448 resulted in a significant decrease in E2F3 mRNA and protein, whereas antimiR-448 showed a positive impact on E2F3 expression. Importantly In silico analysis showed that miR-448 can bind to the 3’UTR region of E2F3 mRNA, we then designed mutations in miR-448seed region binding site in the E2F3 3’UTR, and construct either WT-E2F3-3’UTR or MUT-E2F3-3’UTR luciferase reporter (Figure 6C). We found that miR-448 inhibited, while antimiR-448 promoted luciferase activity of WT luciferase reporter, no significant changes were detected in MUT reporter transfected cells (Figure 6D).
Overexpression of E2F3 reversed the growth inhibition and pro-apoptotic effects of miR-448
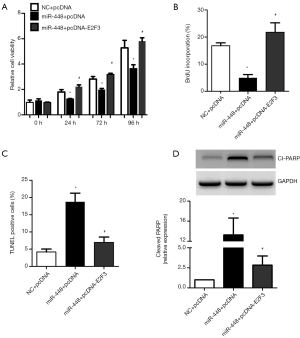
To examine the role of E2F3 in miR-448 mediated growth inhibition and apoptosis, we constructed a plasmid to overexpress E2F3. Overexpression of E2F3 abolished the inhibitory effects of miR-448 on cell proliferation (Figure 7A,B). In addition, overexpression of E2F3 also diminished apoptosis induced by miR-448 (Figure 7C,D). These data suggest that E2F3 is a functional target of miR-448, and may partially mediate the growth inhibition and pro-apoptotic effects of miR-448.
Discussion
In the current study, we demonstrate that a novel tumor suppressive microRNA, miR-448, was constantly downregulated in CRC. Functional study revealed that miR-448 restrained cell growth via inhibiting cell proliferation and inducing apoptotic cell death. Moreover, we show that miR-448 targets the 3’UTR of the mRNA of transcriptional factor E3F3, which is involved in the oncogenesis process. Consistent with the in vitro analysis, we found that E2F3 was upregulated in CRC tissue samples, and a negative correlation between miR-448 and E2F3 expression in these samples was detected. Further we observed that overexpression of E2F3 significantly decreased cell proliferation and diminished the proapoptotic effect by miR-448 administration, suggesting that repression of E2F3 may underlie the anti-proliferation effect of miR-448. Our study therefore identified miR-448 as a key player in CRC and uncovered a novel miR-448/E2F3 signaling pathway for the first time.
MicroRNAs have emerged as one of the key regulators in the development and progression of CRC. Recent studies unraveled several mechanisms of microRNAs guided tumorigenesis in colon. For example, the mediator of inflammatory signaling, STAT3 controls a subset of microRNAs such as miR-21, miR-126 and miR-146, which subsequently activate or suppress NF-κB signaling (6). Moreover, several microRNAs such as miR-34a and miR-339-5p modulates cancer cell survival involving mechanisms associated with p53 signaling (11,12). MiR-448 has been previously shown to be downregulated in gastric cancers and ovarian cancers, ectopic expression of miR-448 exerted antitumor action by suppression of cell proliferation and invasion (9,10). In our study, we first showed that miR-448 was also downregulated in CRC, suggesting a similar anti-tumor role of miR-448 in the bowel. Indeed, our functional tests are in line with the previous studies; moreover, we found that ectopic expression of miR-448 induced apoptosis in CRC cancer cells, which reconfirmed the previous works and expanded our knowledge on this microRNA.
The canonical way of microRNAs to regulate gene expression is through binding with the 3’UTR of a given mRNA, which results in mRNA degradation or translation blockage (3). To understand which gene is directly controlled by miR-448, we utilized in silico analysis and found multiple potential target genes, one of which is E2F3. Experimental studies confirmed this prediction. Moreover, transfection of miR-448 decreased E2F3 at the mRNA level and the protein level, indicating that miR-448 binding directs a targeted E2F3 mRNA degradation process. E2F3 regulates the expression of genes in cell cycle regulation, apoptosis and differentiation, which supported its pivotal role in the control of cell growth (13). Upregulation of E2F3 is commonly seen in a number of cancers (14,15). Clinically, upregulation of E2F3 in hepatocellular carcinoma and prostate cancer is associated with poor prognosis (15,16). Despite the reports showing the frequent activation of E2F3 in cancers, the regulatory mechanisms of E2F3 in cancers, especially in CRC, were less reported. Recent studies demonstrate that E2F3 serves as a target of several microRNAs (17-19). Particularly, Karaayvaz et al. showed that miR-129 targeted E2F3 to enhance the chemosensitivity of CRC cells against 5-fluorouracil treatment (20). Chang et al. showed that miR-503 induced E2F3 repression results in decreased proliferation of SW480 cells (21). Our identification, which showed that miR-448 serves to directly down-regulate E2F3, yielded similar results with the previous investigations. Our findings together with others’ properly exemplified that one gene can be targeted by multiple microRNAs simultaneously.
How miR-448 is been regulated is seldom reported. Intriguingly, Li et al reported recently that miR-448 transcription is repressed by active NF-κB signaling in breast cancer cells, which facilitates epithelia-mesenchyme transition of cancer cells (22). This finding is reminiscent of the decreased miR-448 expression in CRC since NF-KB mediated inflammatory signal is constantly activated in inflammatory bowel diseases and CRC (23), suggesting that miR-448 might play a critical role in the development of CRC. More detailed regulatory mechanism of miR-448 in CRC may be characterized in future. Furthermore, an in vivo study is needed to confirm the miR-448/E2F3 anti-tumor signaling uncovered by the present study.
Collectively, we report for the first time that miR-448 is constantly downregulated in CRC tissues, it produced prominent anti-tumor function in vitro by inhibiting proliferation and inducing apoptosis. Importantly, E2F3 was a target of miR-448 and partially accounts for these tumor suppressive effects. Our findings will help to expand the knowledge of microRNA regulated tumorigenesis and suggest that miR-448 replacement therapy might be effective in combating CRC.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.46). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the affiliated hospital of Qingdao university (approval NO. QYFYEC 2016-012-01) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Rabeneck L, Horton S, Zauber AG, Earle C. Colorectal Cancer. In: Gelband H, Jha P, Sankaranarayanan R, et al. editors. Cancer: Disease Control Priorities. Third Edition (Volume 3). Washington (DC): The World Bank, 2015.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Wang J, Song YX, Ma B, et al. Regulatory Roles of Non-Coding RNAs in Colorectal Cancer. Int J Mol Sci 2015;16:19886-919. [Crossref] [PubMed]
- Li M, Izpisua Belmonte JC. Roles for noncoding RNAs in cell-fate determination and regeneration. Nat Struct Mol Biol 2015;22:2-4. [Crossref] [PubMed]
- Chi Y, Zhou D. MicroRNAs in colorectal carcinoma--from pathogenesis to therapy. J Exp Clin Cancer Res 2016;35:43. [Crossref] [PubMed]
- Shi C, Yang Y, Xia Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 2016;65:1470-81. [Crossref] [PubMed]
- Rokavec M, Öner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest 2014;124:1853-67. [Crossref] [PubMed]
- Wu X, Tang H, Liu G, et al. miR-448 suppressed gastric cancer proliferation and invasion by regulating ADAM10. Tumour Biol 2016;37:10545-51. [Crossref] [PubMed]
- Lv Y, Lei Y, Hu Y, et al. miR-448 negatively regulates ovarian cancer cell growth and metastasis by targeting CXCL12. Clin Transl Oncol 2015;17:903-9. [Crossref] [PubMed]
- Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A 2007;104:15472-7. [Crossref] [PubMed]
- Zhang C, Liu J, Wang X, et al. MicroRNA-339-5p inhibits colorectal tumorigenesis through regulation of the MDM2/p53 signaling. Oncotarget 2014;5:9106-17. [Crossref] [PubMed]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet 2001;10:699-703. [Crossref] [PubMed]
- Olsson AY, Feber A, Edwards S, et al. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene 2007;26:1028-37. [Crossref] [PubMed]
- Foster CS, Falconer A, Dodson AR, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene 2004;23:5871-9. [Crossref] [PubMed]
- Zeng X, Yin F, Liu X, et al. Upregulation of E2F transcription factor 3 is associated with poor prognosis in hepatocellular carcinoma. Oncol Rep 2014;31:1139-46. [PubMed]
- Chen L, Kong G, Zhang C, et al. MicroRNA-432 functions as a tumor suppressor gene through targeting E2F3 and AXL in lung adenocarcinoma. Oncotarget 2016;7:20041-53. [PubMed]
- Yang Y, Chang S, Zhao Z, et al. MicroRNA-214 suppresses the proliferation of human hepatocellular carcinoma cells by targeting E2F3. Oncol Lett 2015;10:3779-84. [PubMed]
- Dong D, Gong Y, Zhang D, et al. miR-874 suppresses the proliferation and metastasis of osteosarcoma by targeting E2F3. Tumour Biol 2016;37:6447-55. [Crossref] [PubMed]
- Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis 2013;4:e659 [Crossref] [PubMed]
- Chang SW, Yue J, Wang BC, et al. miR-503 inhibits cell proliferation and induces apoptosis in colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol 2015;8:12853-60. [PubMed]
- Li QQ, Chen ZQ, Cao XX, et al. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial-mesenchymal transition of breast cancer cells. Cell Death Differ 2011;18:16-25. [Crossref] [PubMed]
- Sun XF, Zhang H. NFKB and NFKBI polymorphisms in relation to susceptibility of tumour and other diseases. Histol Histopathol 2007;22:1387-98. [PubMed]


