
The hypermethylation of MUC2 promoter associated with mRNA and metastasis in esophageal squamous cell carcinoma
Introduction
Esophageal cancer is divided into two main types: squamous cell carcinoma and adenocarcinoma. Esophageal squamous cell carcinoma (ESCC) in China is a type of high incidence of esophageal cancer (1). Despite improvements in clinical treatments, the 5-year survival are still very poor regardless of race and gender and are not significantly improved over the past several decades (2,3).
Epithelial surfaces are protected by mucins, which are secreted by the gastrointestinal tract. As a secretory mucin gene, MUC2 is high expressed in the cells of human colon and small intestine, by which to protect gastro-intestinal tract from damaging by several risk factors (4-6). The abnormal level of MUC2 mRNA and protein has been found in many types of cancers (7). The results found that MUC2 and MUC5AC were decreased in endometrial and cervical tissues (8). Genetic variations in MUC2 could be an important factor for gastric cancer with H. pylori infected patients (9). The expression of MUC2 is decreased in gastric signet-ring cell carcinoma (10). The MUC2 expression was increased induced Bile acids in colon carcinoma cells through involving of AP-1 (11). Galectin-3 could up-regulate of MUC2 transcription through AP-1 activation in colon cancer (12).
The level of the MUC2 mRNA could be controlled by the promoter methylation in tumorigenesis. The expression of MUC2 mRNA is associated with promoter methylation status in mucinous gastric carcinoma (13). The promoter of MUC2 gene was highly average 87% methylated in PANC1 cell line, while average 43% methylated in the BxPC3 cell line and average 33% methylated in normal colon crypts (14). Yokoyama et al. found MUC2-negative expression in pancreatic ductal adenocarcinomas, MUC2(+) expression in intestinal-type intraductal papillary mucinous neoplasms, and MUC2(−) expression in gastric-type intraductal papillary mucinous neoplasms. The promoter methylation of MUC1, MUC2 and MUC4 was significant associated with mucin expression in pancreatic neoplasms (15). Meanwhile, the CpG island methylation MUC2, MUC5AC and MUC6 had a positive association with expression and other clinicopathologic variables (16). The results have found that the mRNA level of MUC2 gene silenced by promoter hypermethylation is a risk factor for a unfavorable clinical outcome in HCC (17). These results indicated that the promoter methylation of MUC2 gene could be an important marker in ESCC.
However, we have no plenty information in the regulation of MUC2 gene expression in ESCC, especially in DNA methylation. The aim is to examine the promoter methylation associated with MUC2 mRNA expression in 310 ESCC patients in our study.
Methods
Patients and tissue samples
The 310 patients were randomly selected from Changzhou Cancer Hospital and Nanyang Center Hospital between 2001 and 2014 in China. The tissues included tumors tissues and the corresponding non-tumor tissues were obtained from surgical resection after operation. The all of the relevant medical recorded were reviewed carefully by three pathologists individually. The patients ageing from 36 to 82 years included 180 men and 130 women. The study was agreed with the ethics standards of the committee on Human Experimentation of the Soochow University.
Methylation analysis of MUC2
We detected the promoter methylation of MUC2 gene by methylation-specific PCR assay as previously described (17). The first was to obtained DNA from ESCC tissues, then the DNA was treated by bisulfite modification according to our previous study (17). MUC2 methylation was measured using primers: forward primer of unmethylated MUC2 gene, 5'-GTTGTTTTATTTTGAAGAAGGTTGTG-3', reverse primer, 5'-TAACAAAAACAATATAAATTACACCCAAA-3'; forward primer of methylated MUC2 gene, 5'-GTTGTTTTATTTTGAAGAAGGTTGC-3', reverse primer, 5'-CGATATAAATTACGCCCGAA-3'. The MSP products were separated by 2% agarose gel electrophoresis and visualized by BIO-RAD Gel Doc XR + (Bio-Rad Laboratories, Inc.). Gels intensity profiles analysis was measured by Quantity One (Bio-Rad Laboratories, Inc.). Quantitative comparison of the band between samples was made for the same gel. We calculated the promoter methylation index (MI) of MUC2 gene by the following formula: 100 × methylated reaction/(unmethylated reaction + methylated reaction). ΔMI defined as MIESCC − MINon-tumor.
MUC2 mRNA expression by QPCR
Total RNA was isolated from 310 ESCC and non-tumor tissues. The first-strand cDNA was synthesized from 2 µg of total RNA. The level of MUC2 mRNA were detected by primers: forward primer (5'-CTTCGACGGACTCTACTACAGC-3') and reverse primer (5'-CTTTGGTGTTGTTGCCAAAC-3') (17). qPCR was carried out by the Mx3000P QPCR System (Stratagene, USA). The qPCR for MUC2 mRNA carried out in following conditions: 95 °C, 30 s; 60 °C, 30 s; and 72 °C, 1 min by 40 cycles. Meanwhile, β-actin mRNA was detected as an internal control. All results were normalized to β-actin.
Statistical analysis
Fisher’s exact test, Chi-square test, and two-sample t-test were used to evaluate the statistical differences among the groups with different clinicopathological data. The survival data were analyzed using the Kaplan-Meier method, differences were determined using the log-rank test. All data was performed using SPSS software 18.0 (SPSS Incorporated, Chicago, IL, USA). P<0.05 was considered significant.
Results
The methylation index (MI) MUC2 gene in ESCC patients
We analyzed the promoter methylation index of MUC2 gene by MSP in ESCC patients. The MI means the methylation index. Our results indicated that the mean MI of MUC2 promoter was 0.68 (95% CI, 0.65–0.70) in tumor tissues, and 0.45 (95% CI, 0.42–0.47) in Non-tumor tissues. This results implied that MI of MUC2 gene was significantly difference between the ESCC tissues and non-tumor tissues (P<0.0001) (Figure 1). The MI of MUC2 promoter was elevated (∆MI>0) in 211 (68.06%) of the 310 ESCC patients but only decreased (∆MI≤0) in 99 (31.94%) of the patients. It could indicate the hypermethylation of MUC2 gene is an important factor for the development of ESCC.
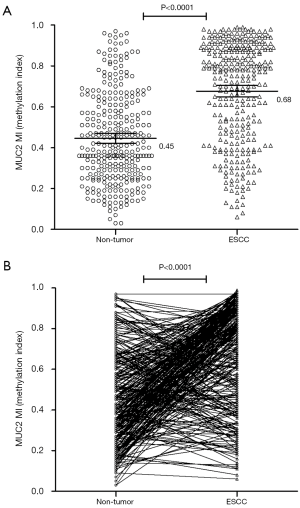
The methylation level of MUC2 gene is associated with and clinical characteristics
The ESCC patients were divided into two groups: MUC2 hypermethylation group (ΔMI>0), 211 cases; MUC2 hypomethylation group (ΔMI≤0), 99 cases, respectively. We compared the related clinical characteristics in two groups of ESCC patients (Table 1). We found a statistical difference between ΔMI≤0 and ΔMI>0 groups in Lymph node metastasis and Distant metastasis; P<0.0001, P=0.026, respectively), by which implied that MUC2 gene hypermethylation could be more in ESCC patients with metastasis cases.
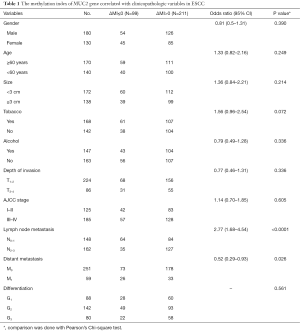
Full table
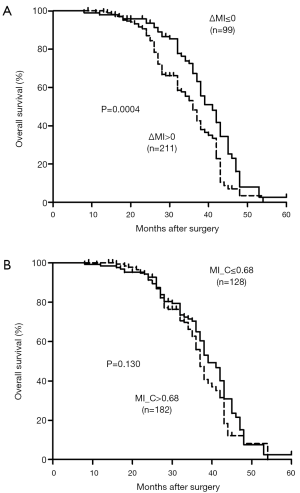
For the next, we want to know the relation between the promoter MI of MUC2 gene and outcomes after surgery, we analyzed the survival by the Kaplan-Meier method in different groups (Figure 2). We found the median cumulative survival was 36 months in hypermethylation cases, but was for 41 months in hypomethylation group (log-rank P=0.0004; HR =1.778; 95% CI, 1.29–2.45). The cutoff value 0.68 was set according to the mean MI of MUC2 in tumor tissues. Meanwhile, we did not find difference in ESCC patient with MI>0.68 (37 months) compared with 39 months in those with MI≤0.68 group (log-rank P=0.130; HR =1.288; 95% CI, 0.93–1.79). These results suggested that ESCC with demethylation from non-tumor to tumor could have a poorer prognostic.
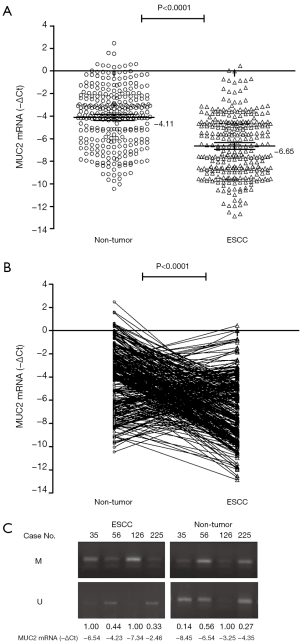
MUC2 mRNA expression in tumor and non-tumor tissues
To quantify relatively MUC2 mRNA levels in tumor and non-tumor tissues by a real-time PCR assay. The expression of MUC2 mRNA is lower in ESCC samples (mean−∆Ct=−6.65; 95% CI, −6.95 to −6.35) than that in non-tumor tissues (mean−∆Ct=−4.11; 95% CI, −4.39 to −3.82). There was a significant difference in MUC2 mRNA between different tissues (P<0.0001, Figure 3). And, we found 104 (33.55%) of the ESCC patients with MUC2 mRNA elevated (−∆∆Ct>0), 206 (66.45%) of the patients with MUC2 mRNA decreased (−∆∆Ct≤0), by which implied MUC2 mRNA silenced could be an important factor in ESCC.
MUC2 mRNA expression associated with clinicopathologic data in ESCC
The expression of MUC2 mRNA in 310 ESCC patients related with clinicopathological variables is summarized in Table 2. There was a statistical difference between MUC2 mRNA with AJCC stage, lymph node metastasis, and differentiation (P=0.029, P=0.001, P<0.0001, respectively). The lower MUC2 mRNA (−ΔΔCtMUC2≤−2.54) was 112/185 (60.54%) in ESCC with AJCC stage III-IV, 105/162 (64.81%) with N2–3 lymph node metastasis, and 99/142 (69.72%) with differentiation G2.
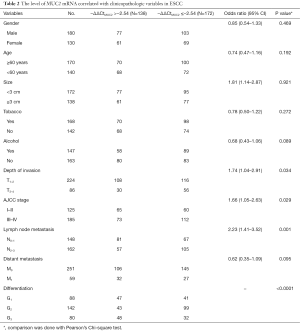
Full table
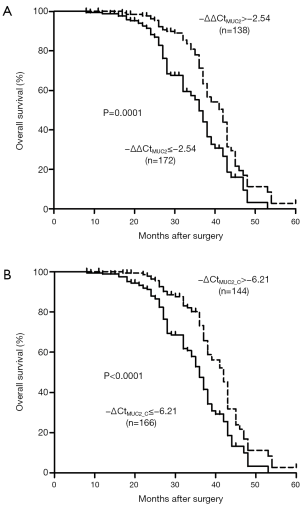
The next, we analyzed the association between MUC2 mRNA and survival of these patients (Figure 4). The resulted indicated that those patients with decreased MUC2 mRNA (−∆∆Ct<−2.54) was significantly poor overall survival (36 vs. 42 months, P=0.0001; HR =0.526; 95% CI, 0.38–0.73). Meanwhile, we also found that ESCC patient with −∆CtMUC2≤−6.21 had a shorter median cumulative survival (36 months) compared with 42 months in those with −∆CtMUC2>−6.21 group (log-rank P<0.0001; HR =0.492; 95% CI, 0.35–0.69). The results gave us information that MUC2 silencing could be an important factor for poor survival in ESCC.
MUC2 mRNA silencing and promoter hypermethylation in ESCC
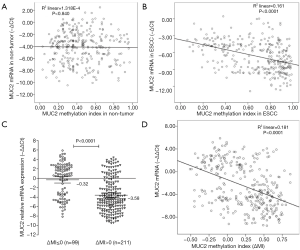
The next, we detected the relation between MUC2 MI and the level of MUC2 mRNA in ESCC tissues (Figure 5). The results indicated ESCC tissues with ΔMI>0 have a lower MUC2 mRNA level (Mean−∆∆Ct=−3.58; 95% CI, −4.07 to −3.09), and ESCC tissues with ΔMI<=0 have a higher MUC2 mRNA level (Mean−∆∆Ct=−0.32; 95% CI, −1.03 to 0.39; P<0.0001; Figure 5C). The decreased MUC2 expression was significant differently in different MI groups of MUC2 gene (R2 =0.161, P<0.0001; Figure 5B; R2 =0.181, P<0.0001; Figure 5D). But, there was no significant association between the demethylation status of MUC2 and mRNA expression in non-tumor tissues (R2 =0.00013; P=0.840; Figure 5A). Thus, the DNA methylation of MUC2 could give us more information for loss of MUC2 mRNA.
Discussion
Mucins have a role for protecting the epithelial surfaces of the related tracts in the human body, which classified as an important factor for some putative transmembrane region (7). The abnormal MUC expression has been implicated in several cancers (18-20). However, Mucins had crucial roles in prognostic prediction and tumor invasion. The MUC2 (+) “intestinal” phenotype was associated with significantly worse prognosis in submucosal gastroesophageal adenocarcinomas, independent of node stage and other prognostic factors (21).
Our previous results indicated that the decreased mRNA expression of MUC2 gene was associated with MUC2 methylation in HCC (17). Research suggested that the DNA methylation could be a biomarker in carcinogenesis (22,23). We found that the mean MI of MUC2 promoter was 0.68 in ESCC, and 0.45 in Non-tumor samples. Also, it indicated a significantly association between the MUC2 MI and distant metastasis, Lymph node metastasis in ESCC patients. The ESCC patients with MUC2 hypomethylation tended to show better survival. Meanwhile, our results found more MUC2 silenced in ESCC tissues, and only 31.94% of ESCC patients with MUC2 mRNA elevated. And a statistical difference was found between MUC2 mRNA with AJCC stage, Lymph node metastasis, and differentiation. Another, ESCC patients with hypermethylation was associated with MUC2 silenced, by which implied that promoter methylation of MUC2 gene could play an important role in ESCC.
The ESCC patients with loss of MUC2 and promoter hypermethylation could be with an unfavorable outcome. There was a significant correlation found between MUC2 mRNA and promoter methylation in ESCC. The key point to methylation alterations could be further investigation the related epigenetic molecular, which is critical for the epigenetic control of gene expression, such as DNA binding and protein-protein interactions in promoter key sites. The next step should give a detail description for these regulatory effects. DNA epigenetic modification could influence MUC2 gene expression in ESCC. It will be interesting to test whether methylation and protein alterations are reversible modifications in future.
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Jiangsu (Grant No.: BL2013012, BE2016656); Changzhou Sci&Tech Program, China (Grant No.: CE20155043, CJ20159023); the Science and Technology Planning Project of Changzhou Health Bureau (Grant No.: ZD201301), the High-level Health Talents of Changzhou City (2016CZLJ021, 2016CZLJ009).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.44). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study had been approved by the Research and Ethical Committee of Changzhou Cancer Hospital of Soochow University (No. 2014006) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shang L, Wang M. Molecular alterations and clinical relevance in esophageal squamous cell carcinoma. Front Med 2013;7:401-10. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Ling Y, Chen J, Tao M, et al. A pilot study of nimotuzumab combined with cisplatin and 5-FU in patients with advanced esophageal squamous cell carcinoma. J Thorac Dis 2012;4:58-62. [PubMed]
- Allen A, Hutton DA, Pearson JP. The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biol 1998;30:797-801. [Crossref] [PubMed]
- Gum JR Jr, Hicks JW, Toribara NW, et al. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J Biol Chem 1994;269:2440-6. [PubMed]
- Tytgat KM, Büller HA, Opdam FJ, et al. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology 1994;107:1352-63. [Crossref] [PubMed]
- Corfield AP. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta 2015;1850:236-52.
- Hebbar V, Damera G, Sachdev GP. Differential expression of MUC genes in endometrial and cervical tissues and tumors. BMC Cancer 2005;5:124. [Crossref] [PubMed]
- Marín F, Bonet C, Muñoz X, et al. Genetic variation in MUC1, MUC2 and MUC6 genes and evolution of gastric cancer precursor lesions in a long-term follow-up in a high-risk area in Spain. Carcinogenesis. 2012;33:1072-80. [Crossref] [PubMed]
- Terada T. An immunohistochemical study of primary signet-ring cell carcinoma of the stomach and colorectum: II. Expression of MUC1, MUC2, MUC5AC, and MUC6 in normal mucosa and in 42 cases. Int J Clin Exp Pathol 2013;6:613-21. [PubMed]
- Song S, Byrd JC, Koo JS, et al. Bile acids induce MUC2 overexpression in human colon carcinoma cells. Cancer 2005;103:1606-14. [Crossref] [PubMed]
- Song S, Byrd JC, Mazurek N, et al. Galectin-3 modulates MUC2 mucin expression in human colon cancer cells at the level of transcription via AP-1 activation. Gastroenterology 2005;129:1581-91. [Crossref] [PubMed]
- Mesquita P, Peixoto AJ, Seruca R, et al. Role of site-specific promoter hypomethylation in aberrant MUC2 mucin expression in mucinous gastric carcinomas. Cancer Lett 2003;189:129-36. [Crossref] [PubMed]
- Hamada T, Goto M, Tsutsumida H, et al. Mapping of the methylation pattern of the MUC2 promoter in pancreatic cancer cell lines, using bisulfite genomic sequencing. Cancer Lett 2005;227:175-84. [Crossref] [PubMed]
- Yokoyama S, Kitamoto S, Higashi M, et al. Diagnosis of pancreatic neoplasms using a novel method of DNA methylation analysis of mucin expression in pancreatic juice. PLoS One 2014;9:e93760 [Crossref] [PubMed]
- Walsh MD, Clendenning M, Williamson E, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol 2013;26:1642-56. [Crossref] [PubMed]
- Ling Y, Zhu J, Gao L, et al. The silence of MUC2 mRNA induced by promoter hypermethylation associated with HBV in Hepatocellular Carcinoma. BMC Med Genet 2013;14:14. [Crossref] [PubMed]
- Cozzi PJ, Wang J, Delprado W, et al. MUC1, MUC2, MUC4, MUC5AC and MUC6 expression in the progression of prostate cancer. Clin Exp Metastasis 2005;22:565-73. [Crossref] [PubMed]
- Boltin D, Niv Y. Mucins in Gastric Cancer - An Update. J Gastrointest Dig Syst 2013;3:15519. [Crossref] [PubMed]
- Elzagheid A, Emaetig F, Buhmeida A, et al. Loss of MUC2 expression predicts disease recurrence and poor outcome in colorectal carcinoma. Tumour Biol 2013;34:621-8. [Crossref] [PubMed]
- Davison JM, Ellis ST, Foxwell TJ, et al. MUC2 expression is an adverse prognostic factor in superficial gastroesophageal adenocarcinomas. Hum Pathol 2014;45:540-8. [Crossref] [PubMed]
- Ahrens TD, Werner M, Lassmann S. Epigenetics in esophageal cancers. Cell Tissue Res 2014;356:643-55. [Crossref] [PubMed]
- Mummaneni P, Shord SS. Epigenetics and oncology. Pharmacotherapy 2014;34:495-505. [Crossref] [PubMed]


