
The challenge for precision medicine: all tumor genomes are different and all cancer patients are different in their own way
Cancer is currently viewed as a disease of evolving genomic instability and abnormal epigenomic modifications. Pioneering work had discovered and chromosomally mapped the genomic locations of oncogenes and tumor suppressor genes that are responsible for cancer initiation, progression and metastasis. By the time the feasibility of the Human Genome Project was being discussed in the early 1980s, cancer research was already facing two options: “either to try to discover the genes important in malignancy by a piecemeal approach or to sequence the whole genome of a selected animal species” (1). The complete human genome sequence was quickly seen to be the necessary foundation for understanding cancer and it became an important driver for the Human Genome Project. Subsequently, the Cancer Genome Project was initiated to categorize the somatic mutations in cancer and it began to reveal that more than 1% of all human genes are implicated in cancer (2). The Cancer Genome Atlas research network has examined a variety of cancers with genetic, transcriptomic, epigenomic and protein expression variants (http://cancergenome.nih.gov/abouttcga). Now, the International Cancer Genome Consortium plans to generate comprehensive catalogues of genomic abnormalities, including somatic mutations, aberrant expression of genes, and epigenetic modifications, in tumors from 50 different cancer types and subtypes from worldwide sources (http://icgc.org/#about). From the mountains of genomic data, we are learning more complete details of the mutation processes that give rise to the common genomic aberrations and the unique genomic expression profiles in specific types of cancer. Advances in sequencing technology and dramatic decreases in its cost are providing us with the potential to accurately inspect the cancer genome at the level of single cells and the resolution of a single nucleotide (3).
The recent paper by Hu et al. (4) is an excellent example of inspecting and summarizing the genome of whole tumors. The authors sequenced the tumor genomes, relative to matched blood DNA, from 15 patients with either gastric cancers (GCA and GNCA) or esophageal squamous cell carcinoma (ESCC) and from each patient, they described the overall genomic landscapes of somatic alterations in single nucleotide variations (SNVs), large genomic structural variations (SVs) and copy number alterations (CNVs). Circos plots provided a summary “snapshot” of the identified genomic alterations from each patient and we will comment on these illustrations of a dramatic degree of genomic heterogeneity, not only between the gastric adenocarcinomas and ESCCs but also within each type of cancer. We can infer that, up to the time each tumor was taken for analysis, there had been a diverse as well as divergent remodeling of the genome during the course of tumor transformation in each tumor and a potential for chaos in the regulation of the cancer genome that underlies an uncontrolled proliferative phenotype. Genome-wide characterization of SNVs revealed that the highest mutation rates were in intergenic regions, followed by introns and exons, and the lowest rates were intragenic mutations, such as missense and nonsense mutations. The lower mutation rates and reduced genetic diversity in coding regions, relative to non-coding regions, suggests there may have been a neo-Darwinian-type selection and adaptation that was occurring during evolution of the tumors.
In the past, we usually ignored the non-coding regions of the genome, but at least 98% of our DNA does not translate into proteins. This previous “dark matter” of the genome does have significant regulatory function, such as found for the long noncoding RNAs (5). Therefore, mutations in the intergenic regions and introns may affect the binding of microRNAs and, subsequently, influence the expression of coding genes (6).
Hu et al. also reported other interesting genomic findings. First, A>C mutations were common in the gastro-esophageal cancers. In addition to the expected 5 prime A, reported in other studies, they also found enrichment of five prime T. Second, the mutational signatures are different in the two types of cancer (4). In gastro-esophageal cancer, enrichment of A>C mutations with prime T or A is common, suggesting there was oxidation of guanine by a potential mutagen. It is becoming clear and, perhaps expected, that different cancers will have unique mutational signatures (7). In lung cancers, C>A transversion often occurs in patients who are smokers, but C>T transition is the predominant type of mutation in non-smokers or previous smokers who have ceased smoking. It is also worth noting that over activity of members of the APOBEC family of cytidine deaminases may play a novel role in endogenous mutagenesis and the generation of intratumor genomic heterogeneity and, as such, it may become an attractive target for individualized therapy (8).
The authors then focused on specific cancer driver mutations and recurrent SVs with a frequency of >20% and 33%, respectively. Their analysis revealed well-known cancer gene mutations in both gastric cancer and ESCC, including TP53, JAK3, BRCA2, FGF2, FBXW7, MSH3, PTCH, NF1, ERBB2, and CHEK2, and potentially novel cancer-associated genes, such as KISS1R, AMH, MNX1, WNK2, and PRKRIR. Those mutated genes involve the interacting or cross-talking DNA repair and tyrosine kinase pathways, which are also common in other cancers. The cluster genomic region deletions (MACAOD2, FHIT, PARK2) were verified in a TCGA-stomach cancer subset. Such SVs are only identified with whole genome sequencing and they could be novel genomic markers.
Although the sample size of patients is small in this study, the findings provide insights into an understanding of the overall changes in tumor genomes and they encourage us to undertake a deeper and more dynamic analysis of tumor evolution. We suggest that our questions should be directed to how can we paint the whole picture of the evolving cancer genome in individual patients and how can we use that information to remedy and manage those individual patients. We are learning the following statements from the present study by Hu et al. (4) and from many similar studies. (I) All normal genomes are the same, but every cancer patient and their cancer genome are different in their own way. The divergent complexity of cancer cells reflects an inevitable genomic and epigenomic heterogeneity and the hallmark of this cancer heterogeneity is the origin of drug resistance, tumor relapse and poor prognosis (we will also imply this in a later discussion of our Figure 1). “Basket trial” driver oncogenic mutation-targeted therapies, which appear to be effective with some common gene mutations, may be a useful consideration (9). The benefits of “basket trial” therapies might be better assessed and customized for precision treatment if, in the future, every cancer genome is sequenced and annotated on an individual basis; however, the evolving genomic and functional heterogeneity within each tumor may present a problem; (II) cancer genome evolution is dynamic in space and time. Most genomic studies of cancer now provide only static information, with a snapshot of the genome at one time point, and hence, they do not give us the whole picture of cancer initiation and its progression. Temporal genomic examination of cell-free tumor DNA is able to monitor the response to treatment of an individual cancer patient and it reflects one of the trends being considered for precision cancer therapy. Nevertheless, the circulating cell-free tumor DNA of an individual patient may not reflect the divergent complexity of intratumor genomic and functional heterogeneity that is evolving in their tumor-cell population and, hence, is contributing to their observed circulating cell-free tumor DNA; (III) knowledge of the genomic changes at different tumor stages may provide not only targets for early diagnosis, but also novel methods for stage-specific treatments and prevention. Defining the actionable mutations and druggable pathways for precision cancer treatment may require not only the cancer genome-sequence from individual cancer patients but also the cancer genome-sequence that is present at specific stages of tumor progression. And last but not least; (IV) the cancer genome is high dimensional with genomic, epigenomic and transcriptomic interactions, and the collective effects of those interactions are more than a simple sum of the parts. Nevertheless, high dimensionality creates the potential for reprogramming the whole system of the cancer genome with small-molecules, chromatin modifiers and immunomodulators; understanding that high dimensionality may reveal effective, individualized strategies to fight cancer. Therefore, we know much about cancer genomics when the elements are collected together, but how should we effectively apply that genomic information is still at the tip of an iceberg and, perhaps many icebergs; a lot still remains to be uncovered and understood before we can approach a cancer cure. Perhaps we should return to some fundamental concepts that underlie the origins of genome architecture (10).
Based on our own and other studies of tumor evolution, we suggest that more efforts should be made to conduct multiple region sampling from individual tumors because they will provide a more dynamic perspective and a greater appreciation for the evolving and divergent complexities of intratumor genomic and functional heterogeneity. Further study of the functional biology of the ontogeny and phylogeny of the intratumor heterogeneity might provide insight into what we are calling the “developmental biology” of tumors in individual patients. If there is to be any potential for successful targeted tumor treatment, we believe that a newer and higher dimensional understanding of tumor biology at the level of evolving individual tumors, including the developmental biology of the evolving and reciprocal interactions between the cellular and micro-environmental complexities of the tumors, will be necessary. Also, we expect that focused collaborative efforts will be required to achieve it.
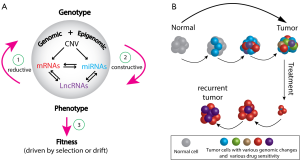
Intratumor genomic heterogeneity was already implicit in the pioneering work by Peter Nowell on the clonal evolution of tumor cell populations (11). More recent, genomic studies have given it a functional reality (12) and they show that we are dealing with the concept of individuality at both the level of the tumor and the individual cancer patient. Since current studies are showing that all tumor genomes are different and all cancer patients are different in their own way, the paper by Hu et al. (4) provides an opportunity to comment on the evolutionary biology and population genetics of tumors and the evolution of the changes in the genomic architecture of tumors (Figure 1). Evolution is a process that is governed by four broad forces that change the genomic variation; it is most easily seen within a sexually reproducing species, but it can be applied to tumor biology. The forces are (I) mutation (including insertions, deletions and duplications); (II) recombination (including gene conversions) within and among chromosomes, and including novel findings of chromothripsis and kataegis in cancer genomes (13); (III) random genetic drift; and (IV) selection. Only selection has the property of adaptation at the level of individual organisms; the other three forces are probabilistic or stochastic events and they are non-adaptive in the sense that they do not depend on the fitness of individual organisms. In effect, cancer could be considered as a new organism.
A biological question is whether individual tumors exhibit an adaptive, selective Neo-Darwinian-type evolution of their cellular genomes or they express a non-Darwinian evolution that is simply the result of chance or random drift of those cellular genomes. We can borrow and modify a discussion from population genetics and organismal biology (Figure 1A) that describes the necessary features for adequacy of a population genetic model for evolutionary change. In genotype space, the language is biochemistry and molecular biology, but in phenotype space, the language is cellular and developmental biology. If different organismal phenotypes have different selective advantages in populations, selection will occur at the level of individuals and their phenotypes, not their genotypes. If there is a genetic basis for selective advantages of different phenotypes, population geneticists have essentially three tasks. One (step 1 in Figure 1A) is to map the visible phenotypic alternates to the genome, a second (step 2 in Figure 1A) is to map those specific genomic alternates to mutually exclusive phenotypes, and a third (step 3 in Figure 1A) is to map those genetically determined, phenotypic alternates to predicted differences in “fitness” of individual organisms. At each step in a population genetic discussion, “context and interaction are of the essence” (14,15). If we can accomplish the three tasks, we have an ability to test whether changes in phenotype of the individuals in a population may be the result of changes in their genotype. A further problem is to return to the central question in population genetics and ask whether the system is driven by a competitive, selective genetic advantage of the individual’s phenotype (such as their survival and contribution of numbers of offspring to the next generation) or is it simply a stochastic process that is driven by chance or random drift.
The evolving divergent complexity of intratumor genomic and functional heterogeneity is depicted in Figure 1B and implies initiation, progression, metastasis, response to drug treatment, and eventual development of therapeutic resistance of tumors. Classical non-Darwinian events of mutation, chromosomal recombination and random drift of the evolving clones of mutant cells may be occurring and driving tumor evolution; however, some mutant cells may be simply adapting and maintaining a stable development in their changing intratumor microenvironment. Hence, the presence of some tumor cells might be the result of a non-Darwinian, clonal expansion and other cells might be the result of selection for their “adaptive fitness” in a specific microenvironment. Multiple micro-samples might provide genomic signatures to distinguish non-Darwinian adaptation from a neo-Darwinian selection by a preponderance of new and novel, nonsynonymous genomic changes that are expected in randomly changing environments relative to synonymous genomic changes that would be expected in more static environments. Contemporary models of punctuated bursts of mutations, chromosomal recombinations (including chromothripsis and kataegis) and random drift of cellular phenotype suggest that non-Darwinian evolution events may be the main forces driving tumor development (16).
In conclusion, we suggest that creative attempts to conduct spatial and temporal analyses of evolving individual tumor genomes should be encouraged. They may be essential analyses to develop novel therapeutic frameworks for precision medicine. The first steps in that direction are the contemporary analyses that are showing intratumor genomic and functional heterogeneity and branching evolutionary processes are hallmarks of cancer. Nevertheless, in those analyses, we are constantly reminding ourselves that the purpose of genomic analysis of cancer is to provide insight, not the cataloging of mutations.
Acknowledgments
Funding: The related work was supported by the National Natural Science Foundation of China (grants 81171992) Henan Scientific and Technological Project (international cooperation grant 152102410088).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Zhen-Yu Lin, MD (Cancer center, Union hospital, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dulbecco R. A turning point in cancer research: sequencing the human genome. Science 1986;231:1055-6. [Crossref] [PubMed]
- Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer 2004;4:177-83. [Crossref] [PubMed]
- Zhang X, Marjani SL, Hu Z, et al. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res 2016;76:1305-12. [Crossref] [PubMed]
- Hu N, Kadota M, Liu H, et al. Genomic Landscape of Somatic Alterations in Esophageal Squamous Cell Carcinoma and Gastric Cancer. Cancer Res 2016;76:1714-23. [Crossref] [PubMed]
- Wu W, Chan JA. Understanding the Role of Long Noncoding RNAs in the Cancer Genome. In: Wu W, Choudhry H. editors. Next Generation Sequencing in Cancer Research-Decoding Cancer Genome. 1st edition. New York: Springer, 2013:199-215.
- Cao W, Wu W, Yan M, et al. Multiple region whole-exome sequencing reveals dramatically evolving intratumor genomic heterogeneity in esophageal squamous cell carcinoma. Oncogenesis 2015;4:e175 [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Swanton C, McGranahan N, Starrett GJ, et al. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov 2015;5:704-12. [Crossref] [PubMed]
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- Lynch M. The Origins of Genome Architechture. Sunderland, MA: Sinauer Associates, Inc., 2007.
- Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23-8. [Crossref] [PubMed]
- Yap TA, Gerlinger M, Futreal PA, et al. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 2012;4:127ps10 [Crossref] [PubMed]
- Kass EM, Moynahan ME, Jasin M. When Genome Maintenance Goes Badly Awry. Mol Cell 2016;62:777-87. [Crossref] [PubMed]
- Lewontin RC. The Genetic Basis of Evolutionary Change. New York: Columbia University Press, 1974.
- Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. New York: Oxford University Press, 1993.
- Ling S, Hu Z, Yang Z, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci U S A 2015;112:E6496-505. [Crossref] [PubMed]


