Differential genomic alterations in normal and cancerous lung cells in response to chronic inflammation
Introduction
Inflammation is a physiological response to tissue damage caused by pathogenic infection, chemical stimulation, and/or wounding (1). The relationship between chronic inflammation and cancer has been widely investigated (2), and numerous studies have demonstrated that chronic inflammation can contribute to carcinogenesis. For example, a study comprising 78,000 patients with asthma demonstrated a high risk of lung cancer in both men and women, consistent with local, chronic inflammation as a contributing factor (3). Previous studies have also reported increased cancer risk among adults with post-inflammatory pulmonary interstitial fibrosis, as seen in patients with silicosis and asbestosis (4). Chronic inflammation may promote some of the molecular changes observed in airway epithelial cells that lead to cancer development (4). Chronic inflammatory disorders appear to increase the risk of cancer development not only in the lung but also in other organs, such as the liver (5), pancreas (6), esophagus (7), and skin (8). In a study involving more than 3,000 patients with chronic ulcerative colitis, a type of inflammatory bowel disease, investigators found an incidence of colon cancer that was 5.7-fold higher than expected (9). Thus, there is increasing evidence supporting the association of chronic inflammation with cancer development.
Chronic inflammation may induce DNA strand breaks, mismatches, and mutations (10), and these events may lead to cellular transformation (2). Genetic alterations such as amplifications and deletions also frequently contribute to carcinogenesis (11). These alterations may change gene expression levels and cause abnormal regulation of the cell cycle and survival pathways. Characterization of these genetic alterations is important for the basic understanding of cancer and its diagnosis. Comparative genomic hybridization (CGH) has been developed to analyze DNA copy number variations (CNVs) across the entire genome. In addition to the CGH assay, there are many other platforms, such as array CGH (aCGH), for analyzing genetic alterations. aCGH can identify DNA CNVs with an emphasis on the most commonly studied genomic coding regions and cancer-related genes (12-14). The advantage of aCGH is its ability to simultaneously detect deletions, duplications, and/or amplifications of any locus represented on an array, with higher resolution than traditional cytogenetic methods. It has also enabled investigators to focus on various types of rearrangements, in particular chromosomal regions. In addition, aCGH is a powerful tool for the detection of submicroscopic chromosomal abnormalities in individuals (15), including subtelomeric and pericentromeric rearrangements (16). Detecting these aberrations by aCGH provides information on the locations of important cancer genes and can have clinical uses for diagnosis, cancer classification, and prognostication (17). In a study of human lung adenocarcinomas, aCGH was used to compare genomic profiles of epidermal growth factor receptor (EGFR) mutant tumors and to identify a proto-oncogene that was specific for an acquired resistance set (18). Another study used aCGH analysis to identify mutations and CNVs that were related to gefitinib sensitivity in non-small cell lung cancers (NSCLC) (19). aCGH can also be used to identify cancer-related prognostic, diagnostic, and therapeutic markers in human cancers induced by chronic inflammation (20). Thus, aCGH provides complete chromosomal information that can reveal genomics-related physiological mechanisms involved in carcinogenesis.
To date, there have been only limited studies of genome-wide screening for chromosomal alterations induced by chronic inflammation. The aim of our study was to identify the CNV regions induced by chronic inflammation in a lung cancer cell line. Normal human lung epithelial cells (BEAS-2B) and cancerous human alveolar epithelial cells (A549) were treated with tumor necrosis factor-alpha (TNF-α). Several molecular mediators, including interleukins, tumor necrosis factor-alpha (TNF-α), and tumor necrosis factor-beta (TGF-β), can promote inflammatory stress and fibrogenic responses in the lung as well as induce progressive, proliferative epithelial reactivity in neighboring cells (21). We used aCGH to identify the CNV regions induced by TNF-α at different time points and found that TNF-α induced more CNV regions in normal lung cells than in cancerous lung cells. Furthermore, we used Ingenuity Pathway Analysis (IPA) to analyze the functions of genes located at the common CNV regions induced by chronic inflammation.
Methods
Cell culture and TNF-α treatment
The normal human lung epithelial cell line BEAS-2B and cancerous human alveolar epithelial cell line A549 were cultured in RPMI 1640 medium supplemented with 1.5 g/L Na2HCO3, 4.5 g/L glucose, and 10% fetal bovine serum (GIBCO-BRL) in a humidified atmosphere containing 5% CO2 at 37 °C. To analyze cell responses under chronic inflammation, the two cell lines were treated with or without 10 ng/mL TNF-α (Alexis Biochemicals, Grünberg, Germany) for 24 weeks. Total RNA and genomic DNA were extracted for analysis at different time points after TNF-α treatment.
Total RNA and genomic DNA extraction
Cells (1.5×106) were washed twice with ice-cold PBS and suspended in 10 mL of ice-cold PBS, followed by centrifugation at 1,000 rpm for 5 min at 4 °C. Total RNA was extracted by TRIzol using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturers’ instructions. Genomic DNA was extracted via a similar protocol using the DNeasy Mini Kit (Qiagen).
Quantitative RT-PCR analysis
Purity of total RNA was confirmed by 1% agarose gel electrophoresis and Bioanalyzer analysis. RNA (1 µg) was reversed transcribed into cDNA using Superscript III (Invitrogen) with random decamer primers (Ambion Europe Ltd., Huntingdon, UK). cDNA transcripts were analyzed using the ABI 7300 Real-Time PCR System. The following primers were used: TNF-α forward 5'-TGGCCCAGGCAGTCAGA-3' and reverse 5'-CAACATGGGCTACAGGCTTGT-3', Interleukin-8 (IL-8) forward 5'-CCAGGAAGAAACCACCGGA-3' and reverse 5'-GAAATCAGGAAGGCTGCCAAG-3', and GAPDH forward 5'-CTAGGCCCCTCCCCTCTTC-3' and reverse 5'-CCCCCACCACACTGAATCTC-3'. All primer oligonucleotides were obtained from Genomics BioSci & Tech Ltd. (Taipei, Taiwan). Each experiment was performed in triplicate. The relative gene copy numbers were calculated by the concentration-CT standard curve method and normalized using the average expression of GAPDH.
aCGH assay
Genomic DNA labeling and hybridization were performed according to the manufacturer’s instructions (Agilent Technologies Inc., Wilmington, DE, USA). In brief, genomic DNA (300 ng) from normal and cancerous lung cells was digested with AluI and RsaI at 37 °C for 2 h. The digested DNA was labeled by random priming for 2 h at 37 °C using the Agilent Genomic DNA Labeling Kit Plus (Agilent Technologies Inc). Genomic DNA from TNF-α-treated cells was labeled with Cy5-dUTP, while that from untreated cells was labeled with Cy3-dUTP. Labeled products were purified using Microcon YM-30 filters (Millipore, Billerica, MA, USA) and then mixed with Human Cot I DNA (Invitrogen). Agilent 105k CGH oligonucleotide microarrays were hybridized with the purified probes at 65 °C for 24 h at 15 rpm, and the slides were then washed.
aCGH data analysis
aCGH arrays were scanned with an Axon scanner and quantified with GenePix software. The raw data were normalized by a procedure utilizing the dependency of neighboring probes, and ridge-tracing normalization, performed in MATLAB (22), was used to adjust the aCGH data. The circular binary segmentation, implemented in the DNAcopy R package, was used to segment the normalized data. Segmentation was used to define the different CNVs in the chromosomes of treated and untreated cells. The absolute values of the log ratios of the regions were compared to those of the derivative log ratio spreads. A region was assigned as a “gain” if the log ratio was positive and as a “loss” if the log ratio was negative. The frequencies of different lengths of identified CNV regions were calculated.
Results
TNF-α induced chronic inflammation
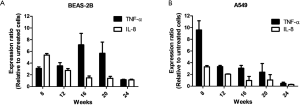
To analyze the degree of inflammation induced by TNF-α, the gene expression levels of two inflammatory marker cytokines, TNF-α and IL-8, were measured in normal and cancerous lung cells treated with TNF-α for 8-24 weeks. The expression levels of TNF-α and IL-8 were 3- and 5.4-fold higher, respectively, in treated BEAS-2B cells than that in untreated BEAS-2B cells. This upregulation of the expression of TNF-α and IL-8 was observed at 12, 16, 20, and 24 weeks (Figure 1A). In A549 cells, the expression of TNF-α was upregulated through 20 weeks, but IL-8 showed no significant upregulation after 12 weeks (Figure 1B). These results indicated that TNF-α induced chronic inflammation in BEAS-2B and A549 cells through at least 20 weeks.
TNF-α induced more CNVs in normal lung cells
To identify CNV regions, aCGH analysis was performed to compare the genomic DNA of both cell lines with or without TNF-α treatment for 16, 20, and 24 weeks. At these respective time points, 261, 319, and 451 CNV regions were identified in TNF-α-treated BEAS-2B cells, whereas TNF-α-treated A549 cells had only 85, 132, and 419 CNV regions (Table 1). This suggested that chronic inflammation by TNF-α induced more CNVs in normal lung cells than in cancerous lung cells.

Full table
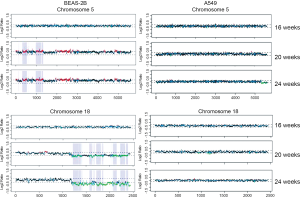
Next, we analyzed the size distribution of the identified regions from the two cell lines at different time points. Most CNV regions induced by TNF-α in BEAS-2B cells were less than 5×105 bp, while only few showed large CNV regions (>6×105 bp). The number of small (1×105-5×105 bp) CNV regions was 234, 143, and 161 at 16, 20, and 24 weeks, respectively (Figure 2A). A similar result was observed in A549 cells (Figure 2B). These results suggested that chronic inflammation induced genomic variations of relatively small size in both TNF-α-treated cells. The location and frequency of CNV regions in each BEAS-2B and A549 chromosome are shown in Figure 3. CNV regions identified as “gain” in BEAS-2B cells are shown in red in Figure 3A, and most were located on chromosomes 5, 9, 11, and 16, whereas CNV regions identified as “losses” are shown in green, and most were located on chromosomes 4, 5, 8, 11, 18, and 19 (Figure 3A). CNV regions were not observed at the same chromosome positions in A549 cells (Figure 3B).


Chronic inflammation induced more genes in common CNV regions in normal cells
The number of accumulated CNV regions were determined in A549 and BEAS-2B cells. There were 46 CNV regions in common at 20 and 24 weeks in A549 cells compared with 984 in BEAS-2B cells. Most of the CNV regions identified as “gain” in BEAS-2B cells were present at 20 and 24 weeks on chromosome 5 (5p12-5p13.3, 5p15.1-5p15.33, 5q14.3; red lines within blue regions in Figure 4), while most of the common “loss” were present on chromosome 18 (18q12.3, 18q21.1-18q21.33, 18q22.1-22.3, 18q23; green lines within blue regions in Figure 4). Very few common CNV regions were identified on chromosome 5 or 18 in A549 cells (Figure 4).
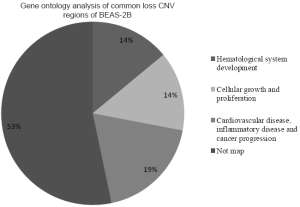
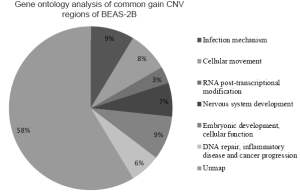
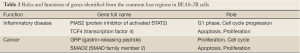
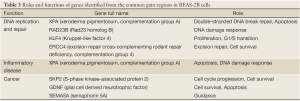
The genes located in the common CNV regions of BEAS-2B cells were identified, and their functions or pathway involvements were analyzed by IPA. Of the 107 genes identified from the common loss regions, nine were involved in hematological system development, nine were related to cellular growth and proliferation, and only four [protein inhibitor of activated STAT2 (PIAS2), transcription factor 4 (TCF4), gastrin-releasing peptide (GRP), and SMAD family member 2 (SMAD2)] were related to functional pathways in inflammatory disease and cancer progression (Table 2; Figure S1, Table S1). The remaining genes located in the common “loss” regions could not be mapped to any functional studies or pathways by IPA. Among the 151 genes identified in the common “gain” CNV regions in BEAS-2B cells, 13 were associated with infection mechanisms, 12 were involved in cellular movement, five were related to post-transcriptional RNA modifications, 10 were related to nervous system development, and 13 were associated with embryonic development and cellular function. There were only seven genes (Figure S2, Table S2), xeroderma pigmentosum, complementation group A (XPA), Rad23 homolog B (RAD23B), Krüppel-like factor 4 (KLF4), excision repair cross-complementing rodent repair deficiency complementation group 4 (ERCC4), glial cell derived neurotrophic factor (GDNF), semaphorin 5A (SEMA5A), and S-phase kinase-associated protein 2 (SKP2) (Table 3), related to DNA replication, DNA repair, inflammatory disease, and cancer progression.
Full table
Full table
Discussion
In this study, normal (BEAS-2B) and cancerous (A549) lung cells were treated with TNF-α to induce inflammation. Real-time PCR analysis of the inflammatory markers IL-8 and TNF-α suggested that TNF-α can lead to chronic inflammation in both BEAS-2B and A549 cells. In addition, during a chronic inflammation state, more CNV regions were found in normal lung cells than in cancerous lung cells. Some of the genes located in these common CNV regions were identified using IPA and may be involved in carcinogenesis and immune response.
Chronic or prolonged inflammation leads to a progressive shift in cells present at the site of inflammation. Cytokines are a class of signaling molecules that are essential for the inflammatory process (23,24). The pro-inflammatory cytokine TNF-α, which participates in the initiation and maintenance of inflammation, regulates inflammatory cells and mediates many inflammatory processes. In this study, TNF-α treatment enhanced mRNA expression level of TNF-α for up to 20 weeks in normal and cancerous lung cells (Figure 1). Furthermore, expression levels of IL-8, considered to be an early-phase inflammatory marker (25,26), were higher in TNF-α-treated cells than in untreated cells before 16 weeks. Therefore, we suggest that TNF-α can induce inflammation for at least 20 weeks in normal and cancerous lung cells. Our results also showed that the expression level of IL-8 decreased, but was still upregulated at 24 weeks in normal TNF-α-treated lung cells compared with its level in untreated cells; however, no change was observed in the expression level of IL-8 at 20 and 24 weeks in the cancerous lung cells. Under inflammatory conditions, cells can synthesize various enzymes and cytokines that are active against inflammation (27). Furthermore, IL-8 is regulated, in part, by anti-inflammatory cytokines to decrease the expression and function of pro-inflammatory cytokine signals (28). Therefore, cells have the potential to auto-regulate their inflammatory status and decrease the expression level of IL-8. The cancerous tumor cells showed no changes in expression levels of TNF-α and IL-8 at weeks 20 and 24, indicating that the cells may have already adapted or become resistant to TNF-α treatment. Taken together, these data may suggest that chronic inflammation can be induced by TNF-α treatment in normal and cancerous lung cells.
Since its development, aCGH has been used routinely in genetic instability studies to characterize DNA polymorphisms, mutations, and CNVs [59,60]. Previous studies have reported that a prolonged inflammatory response can increase DNA mutations and genomic instability (29-31). In an inflammation-associated liver cancer mouse model, Mdr2-KO mice, early inflammatory stages resulted in apoptosis, cell cycle arrest, increased genomic instability, and carcinogenesis (32). Inflammation also induces genomic instability through inflammatory mediators, leading to accumulation of genetic alterations in cells (33). Our results suggest that chronic inflammation induces genomic instability, including increased CNVs that can be detected by aCGH.
In this study, more CNVs were detected in TNF-α-treated BEAS-2B cells than in untreated BEAS-2B cells. In contrast, fewer CNVs were detected in TNF-α-treated A549 cells than in untreated A549 cells (Table 1). Chronic inflammation can create significant environmental stress in cells and tissues, and it has been reported that this stress leads to decreased activity of the DNA repair system, which may lead to more DNA mutations and subsequently enhance genomic instability in other genes (34). Thus, the stress of chronic inflammation may cause more CNVs in normal lung cells by decreasing the activity of the DNA repair system. An earlier study using M-FISH assays reported that A549 cells had many chromosomal alterations, including abnormalities in chromosome numbers and inter-chromosomal translocations, because of repair deficiency (35). Since A549 cells already have many chromosomal alterations, chronic inflammation in TNF-α-treated A549 cells appears to be unable to induce additional genomic instability by decreasing DNA repair activity. In addition, the lower expression level of TNF-α at 20 and 24 weeks in TNF-α-treated A549 cells indicates that these cells may be more resistant to the inflammatory environment, and therefore, induce fewer DNA altered regions compared to that untreated cells. Therefore, our results suggest that chronic inflammation can induce more CNVs in normal than in cancerous lung cells.
Although chronic inflammation can increase genomic instability throughout the treatment period, common CNV regions were only found at 20 and 24 weeks and could not be identified in normal lung cells at earlier time points. A previous study mentioned that genomic instability usually occurs in the early phase of tumor progression (36). Another study showed that random karyotypic changes were only observed in SV40-transformed BEAS-2B cells during the first 12 passages following viral transformation; however, the cells steadily accumulated characteristic abnormal chromosomes corresponding to carcinogenic phenotypes during extended in vitro passages (37,38). Compared to A549 cells, BEAS-2B cells may have still been at an early phase of cancer progression, during which chromosomal alterations could be induced easily (39). Thus, many chromosomal alterations are induced by chronic inflammation, but only some of these may remain and accumulate during cell passages to become the common CNV regions observed at late passages in normal lung cells. In addition, although some studies have reported that the number of gene copies has no effect on relative expression levels in some tissues, a small but significant positive correlation was found here between gene expression levels and CNVs (40). Recent studies also reported that up to 20% of differentially expressed genes were associated with CNVs in humans and rats (41-43). If these accumulated common CNV regions contain genes related to DNA repair or cell cycle regulation, oncogenes, or cancer suppressor genes, alteration of their expression levels by genomic changes could enhance carcinogenesis in normal lung cells. Therefore, it is important to understand the potential functions of the genes located in the accumulated common CNV regions.
Many accumulated common CNV regions were found after chronic inflammation induced by TNF-α; however, only a few of the genes in these common CNV regions were identified to be involved in DNA replication or the DNA repair system. RAD23B, XPA, and ERCC4, which are DNA repair enzymes involved in the nucleotide excision repair (NER) system, were located in the identified common gain regions. RAD23B is an essential protein for NER interaction with damaged DNA resulting in DNA structural changes, specifically opening of the DNA double helix (44-46). It has been observed that dysregulation of RAD23B in dysplastic livers may cause chromosomal instability in TGFα/c-myc double-transgenic mice and enhance hepatocarcinogenesis (47,48). XPA is a DNA repair protein complementing XP-A cells that has been reported to interact with a DNA excision repair-related gene (49). Irradiating XPA-deficient mice with UV light can cause photobiologic reactions including inflammation, genetic instability, and increased epidermal cell carcinogenesis (50). A clinical study found that skin exposure to UV light may increase the risk of cancer development in XP patients (51). In addition to XPA, excision repair cross-complementing (ERCC) genes are also indispensable for NER. The function of ERCC family is to decrease DNA damage by nucleotide excision and repair (52). NSCLC patients with high expression of ERCC1-ERCC4 have better survival than patients with low expression of ERCC1-ERCC4 (52). Based on these previous studies, expression of DNA NER-related genes may contribute to cancer progression both in vivo and in vitro. Therefore, during chronic inflammation, CNVs may dysregulate the activity of genes involved in DNA replication or DNA repair, enhancing genomic alterations and instability and inducing tumor progression.
KLF4, another gene located in the common CNV region, is a regulator of proliferation and differentiation, and altered KLF4 expression has been found in a number of cancers including esophageal and gastrointestinal cancer (53,54). In animal models, KLF4 can activate a number of inflammatory cytokines, such as TNF-α and IL-1α, and can activate inflammation-mediated esophageal squamous cell cancer (55). Semaphorin 5A (Sema5A), an axon regulator, has been associated with carcinogenic processes. Studies have reported that Sema are putative tumor suppressors, anti-angiogenic factors, and mediators of tumor angiogenesis, invasion, and metastasis (56,57). Pan et al. reported that Sema5A may be associated with gastric carcinogenesis (58). In a previous study, the downregulation of Sema5A in cancerous lung tissue, both at the transcriptional and translational levels, was associated with poor survival among nonsmoking women with NSCLC in Taiwan (59). The cancer-related gene, SKP2, is one subunit of the ubiquitin-protein ligase complex, which is also required for the degradation of the tumor suppressor p27. Overexpressed SKP2 has been detected in many different human cancers (60). Abnormal expression of SKP2 is not only a common feature of NSCLC, but also appears to be involved in squamous cell carcinoma progression (61). The expression levels of these genes with CNVs require further validation by real-time PCR. Nevertheless, the findings of our study may suggest that these genes located in the common CNV regions are highly associated with carcinogenesis and that their expression levels may be altered to increase either genomic instability or carcinogenesis in normal lung cells under chronic inflammation.
Conclusions
The data in this study showed that chronic TNF-α treatment induced more CNVs and common CNV regions in normal than in cancerous lung cells. These results suggest that chronic inflammation may cause genomic alterations in genes involved in DNA repair or cancer-related functions, leading to increased carcinogenesis in normal lung cells.
Full table

Full table
Acknowledgments
Funding: This work was supported in part by NSC-95-2314-B-002-112-MY3 from the National Science Council, Taiwan, ROC.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.04.12). EYC serves as the Editor-in-Chief of Translational Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki. The study needs no approval of the Institutional Review Board (IRB) due to containing no samples from patients. Informed consent was obtained.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Philip M, Rowley DA, Schreiber H. Inflammation as a tumor promoter in cancer induction. Semin Cancer Biol 2004;14:433-9. [PubMed]
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860-7. [PubMed]
- Vesterinen E, Pukkala E, Timonen T, et al. Cancer incidence among 78,000 asthmatic patients. Int J Epidemiol 1993;22:976-82. [PubMed]
- Schottenfeld D, Beebe-Dimmer J. Chronic inflammation: a common and important factor in the pathogenesis of neoplasia. CA Cancer J Clin 2006;56:69-83. [PubMed]
- Connolly MK, Bedrosian AS, Mallen-St Clair J, et al. In liver fibrosis, dendritic cells govern hepatic inflammation in mice via TNF-alpha. J Clin Invest 2009;119:3213-25. [PubMed]
- Hsing AW, Sakoda LC, Chua S Jr. Obesity, metabolic syndrome, and prostate cancer. Am J Clin Nutr 2007;86:s843-57. [PubMed]
- Oh DS, DeMeester SR, Vallbohmer D, et al. Reduction of interleukin 8 gene expression in reflux esophagitis and Barrett’s esophagus with antireflux surgery. Arch Surg 2007;142:554-9; discussion 559-60. [PubMed]
- Muller AJ, Sharma MD, Chandler PR, et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc Natl Acad Sci U S A 2008;105:17073-8. [PubMed]
- Ekbom A, Helmick C, Zack M, et al. Ulcerative colitis and colorectal cancer. A population-based study. N Engl J Med 1990;323:1228-33. [PubMed]
- Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res 2001;477:7-21. [PubMed]
- Coleman WB, Tsongalis GJ. Molecular mechanisms of human carcinogenesis. EXS 2006;321-49. [PubMed]
- Kallioniemi A, Kallioniemi OP, Piper J, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A 1994;91:2156-60. [PubMed]
- Iwabuchi H, Sakamoto M, Sakunaga H, et al. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res 1995;55:6172-80. [PubMed]
- Riopel MA, Spellerberg A, Griffin CA, et al. Genetic analysis of ovarian germ cell tumors by comparative genomic hybridization. Cancer Res 1998;58:3105-10. [PubMed]
- Shaffer LG, Bejjani BA, Torchia B, et al. The identification of microdeletion syndromes and other chromosome abnormalities: cytogenetic methods of the past, new technologies for the future. Am J Med Genet C Semin Med Genet 2007;145C:335-45. [PubMed]
- Locke DP, Segraves R, Carbone L, et al. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res 2003;13:347-57. [PubMed]
- Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet 2005;37:S11-7. [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [PubMed]
- Gandhi J, Zhang J, Xie Y, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One 2009;4:e4576 [PubMed]
- Katoh M, Katoh M. Bioinformatics for cancer management in the post-genome era. Technol Cancer Res Treat 2006;5:169-75. [PubMed]
- Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature 2008;454:436-44. [PubMed]
- Chen HI, Hsu FH, Jiang Y, et al. A probe-density-based analysis method for array CGH data: simulation, normalization and centralization. Bioinformatics 2008;24:1749-56. [PubMed]
- Akira S, Isshiki H, Sugita T, et al. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J 1990;9:1897-906. [PubMed]
- Kishimoto T, Hibi M, Murakami M, et al. The molecular biology of interleukin 6 and its receptor. Ciba Found Symp 1992;167:5-16; discussion 16-23. [PubMed]
- Lam HS, Ng PC. Biochemical markers of neonatal sepsis. Pathology 2008;40:141-8. [PubMed]
- Zuppa AA, Calabrese V, D’Andrea V, et al. Evaluation of C reactive protein and others immunologic markers in the diagnosis of neonatal sepsis. Minerva Pediatr 2007;59:267-74. [PubMed]
- Wigmore SJ, Fearon KC, Maingay JP, et al. Interleukin-8 can mediate acute-phase protein production by isolated human hepatocytes. Am J Physiol 1997;273:E720-6. [PubMed]
- de Waal Malefyt R, Abrams J, Bennett B, et al. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med 1991;174:1209-20. [PubMed]
- Federico A, Morgillo F, Tuccillo C, et al. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer 2007;121:2381-6. [PubMed]
- Macarthur M, Hold GL, El-Omar EM. Inflammation and Cancer II. Role of chronic inflammation and cytokine gene polymorphisms in the pathogenesis of gastrointestinal malignancy. Am J Physiol Gastrointest Liver Physiol 2004;286:G515-20. [PubMed]
- Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol 2004;287:G7-17. [PubMed]
- Barash H. Accelerated carcinogenesis following liver regeneration is associated with chronic inflammation-induced double-strand DNA breaks. Proc Natl Acad Sci U S A 2010;107:2207-12. [PubMed]
- Colotta F, Allavena P, Sica A, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 2009;30:1073-81. [PubMed]
- Chang CL, Marra G, Chauhan DP, et al. Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol 2002;283:C148-54. [PubMed]
- Berrieman HK, Ashman JN, Cowen ME, et al. Chromosomal analysis of non-small-cell lung cancer by multicolour fluorescent in situ hybridisation. Br J Cancer 2004;90:900-5. [PubMed]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759-67. [PubMed]
- Loeb LA, Monnat RJ Jr. DNA polymerases and human disease. Nat Rev Genet 2008;9:594-604. [PubMed]
- Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934. [PubMed]
- Ohnuki Y, Reddel RR, Bates SE, et al. Chromosomal changes and progressive tumorigenesis of human bronchial epithelial cell lines. Cancer Genet Cytogenet 1996;92:99-110. [PubMed]
- Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet 2009;18:R1-8. [PubMed]
- Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007;315:848-53. [PubMed]
- Guryev V, Saar K, Adamovic T, et al. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 2008;40:538-45. [PubMed]
- Lu TP, Lai LC, Tsai MH, et al. Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PLoS One 2011;6:e24829 [PubMed]
- Schauber C, Chen L, Tongaonkar P, et al. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 1998;391:715-8. [PubMed]
- Evans E, Moggs JG, Hwang JR, et al. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J 1997;16:6559-73. [PubMed]
- Tapias A, Auriol J, Forget D, et al. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem 2004;279:19074-83. [PubMed]
- Cadet J, Bourdat AG, D’Ham C, et al. Oxidative base damage to DNA: specificity of base excision repair enzymes. Mutat Res 2000;462:121-8. [PubMed]
- Hironaka K, Factor VM, Calvisi DF, et al. Dysregulation of DNA repair pathways in a transforming growth factor alpha/c-myc transgenic mouse model of accelerated hepatocarcinogenesis. Lab Invest 2003;83:643-54. [PubMed]
- Li L, Elledge SJ, Peterson CA, et al. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci U S A 1994;91:5012-6. [PubMed]
- Horio T, Miyauchi-Hashimoto H, Kuwamoto K, et al. Photobiological information obtained from XPA gene-deficient mice. Photochem Photobiol 2007;83:218-24. [PubMed]
- van Steeg H, Kraemer KH. Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol Med Today 1999;5:86-94. [PubMed]
- Simon GR, Sharma S, Cantor A, et al. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 2005;127:978-83. [PubMed]
- McConnell BB, Ghaleb AM, Nandan MO, et al. The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays 2007;29:549-57. [PubMed]
- Chen X, Whitney EM, Gao SY, et al. Transcriptional profiling of Krüppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol 2003;326:665-77. [PubMed]
- Tetreault MP, Wang ML, Yang Y, et al. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology 2010;139:2124-2134.e9.
- Potiron VA, Roche J, Drabkin HA. Semaphorins and their receptors in lung cancer. Cancer Lett 2009;273:1-14. [PubMed]
- Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat Rev Mol Cell Biol 2005;6:789-800. [PubMed]
- Pan G, Lv H, Ren H, et al. Elevated expression of semaphorin 5A in human gastric cancer and its implication in carcinogenesis. Life Sci 2010;86:139-44. [PubMed]
- Lu TP, Tsai MH, Lee JM, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev 2010;19:2590-7. [PubMed]
- Hung WC, Tseng WL, Shiea J, et al. Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Lett 2010;288:156-61. [PubMed]
- Zolota VG, Tzelepi VN, Leotsinidis M, et al. Histologic-type specific role of cell cycle regulators in non-small cell lung carcinoma. J Surg Res 2010;164:256-65. [PubMed]