
USP11 promotes tumorigenesis and enhances MRI detection in breast cancer
Introduction
Breast cancer is one of the most common malignancy in females and the second leading cause of cancer-related mortality all over the world (1,2). In china, metastatic related deaths of breast cancer continuously rise at 3% per year (3). Although more and more biomarkers were reported to act as predictors of malignant status, the early and effective novel therapeutic targets still need further explore. Among numerous the targets, we focus our interest on the Enhancer of zeste homologue 2 (EZH2), which is the enzymatic subunit of the polycomb-repressive complex 2 (PRC2) (4), and mainly work as histone H3 lysine 27 (H3K27) trimethyltransferase using the SET domain (5). More and more groups have reported that EZH2 was aberrantly overexpressed in various types of cancers, such as breast, colon, prostate and liver cancers (6), and the high expression of EZH2 was associated with cancer cell proliferation, migration, and invasion abilities. Especially in breast cancer (7-9), EZH2 was correlated with histological grade, estrogen receptor (ER), as well as progesterone receptor (PR) expression, and might be an independent prognostic factor for OS, DFS and MFS (10,11). But the questions which still disturb us are why EZH2 is highly expressed and the regulation of EZH2 is still uncertain. As we known, the ubiquitination of proteins was an essential step for degradation and was involved in the modification of the target proteins. To the contrary, the antagonizing effect of ubiquitylation was induced by deubiquitylating enzymes (DUBs).
USP11 (ubiquitin-specific protease 11), was a member of the deubiquitylating enzyme families. USP11 was reported to stabilize and modulate the HPV-16E7 (12). Similarly, BRCA2 could be deubiquitinated by USP11 (13) and therefore exhibits its preserving function of DNA damage. The type I TGFb receptor (ALK5) could also be deubiquitinated by USP11, which resulting in enhanced TGFb-induced gene transcription (14). However, knowledges about the biological roles of USP11 in breast cancer progression are still lacking. By far, there was only one paper about that low USP-11 expression was correlated with better survival outcomes (15).
In this study, we identified EZH2 was a substrate for USP11, and the function of USP11 was to promote tumorigenesis, metastasis and enhancing MRI detection in breast cancer. On the whole, we were trying to reveal USP11 worked as a new biomarker and therapeutic target in triggering breast cancer.
Methods
Tumor specimens
Breast cancer tumor specimens and their adjacent normal tissues were received from 120 patients who were diagnosed as breast cancer and underwent breast surgery in our hospital from January 2007 to December 2013. The tissues were obtained after surgery and stored at −80 °C immediately until used for mRNA isolation. The tumor samples were classified according to the American Society of Clinical Oncology (ASCO). Patients subjected to preoperative chemotherapy before surgery were excluded. Written consent was obtained from all participants prior to the study. The ethical approval was given by the hospital ethics committee and the ethics approval ID was Suzhou University-Ethics-D-20150831. The survival times were calculated from operation to recurrence or metastasis related death.
Cell lines
Human breast cancer cell lines MCF-7 cells (luminal), MDA-MB-231 cells (triple-negative) and the normal human mammary epithelial cells (MCF-10A) were purchased from the American Type Culture Collection (Manassas, VA, USA). All the above cells were cultured in DMEM medium or 1640 medium or the MEGM Bullet Kit (HyClone) containing 10% fetal bovine serum (GIBCO, NY, USA) and then supplemented with antibiotics (100 U/mL penicillin and 100 g/mL streptomycin; Sigma-Aldrich, USA). Cells were cultured in a humidified 5% CO2 incubator at 37 °C.
Reagents and antibodies
Lipofectamine 2000 reagent and Lipofectamine RNAiMAX (Invitrogen) were used for plasmids or siRNAs transfection. Cells were transfected at 70% confluent and were routinely passaged at 90–95% confluent, according to manufacturer’s instructions. Matrigel was from BD Biosciences (San Diego, CA, USA). Non-targeting siRNA was used as a negative control (NC). Two different small interfering RNAs (siRNAs) targeting USP11 (siUSP11) or EZH2 (siEZH2) were synthesized respectively (Genechem, Shanghai, China). Antibodies used were as follows: anti-USP11 antibody (Cell Signaling Technology, Danvers, MA, USA), anti-EZH2 antibody (BD Biosciences, Bedford, MA, USA) and anti-β-actin (Sigma-Aldrich). The secondary antibody was purchased from Santa Cruz (Santa Cruz, CA, USA).
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA of cell lysates was isolated using Trizol solution (Invitrogen Life Technologies, Carlsbad, CA, USA), according to the manufacture’s introduction. The quality and concentration of the RNA was measured with a RNA Pico chip. Reverse transcription was performed using 1 µg of total RNA (Invitrogen Life Technologies), Oligo (deoxythymidine)20 was used as primers.
qRT-PCR was performed on an ABI 7900 Fast System Real-Time detection system (Applied Biosystems, Foster City, CA, USA) using SYBR (Roche). GAPDH mRNA was used as an internal normalization control. The amplification protocol was: denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 sec, annealing and extension at 60 °C for 30 sec. All the experiments were performed in three times. Specific primers for the USP11 gene: forward primer CCGTGACTACAACAACTCCT; reverse primer TCGTCATCTTCTTTCTCATCCC.
Co-immunoprecipitation assay
MCF-cells or MDA-MB-231 cells were lysed with cold lysis buffer (50 mM Tris-Cl, 0.5% NP-40, 150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, pH 7.4 and protease inhibitor cocktail) on a rotator at 4 °C for 1 hour. The supernatants of the whole cell lysates were incubated with appropriate primary antibodies such as USP11 or EZH2 or normal rabbit/mouse immunoglobulin G (IgG) on a rotator at 4 °C overnight, and then the immune complexes were added with protein A/G Sepharose CL-4B beads for 2 h at 4 °C. After washed 5 times with lysis buffer, the immune complexes with beads were subjected to SDS-PAGE, followed by western blot analysis.
GST pull down assay
GST-USP11 fusion construct was expressed in BL21 Escherichia coli cells. The in vitro transcription and translation of EZH2 was done using rabbit reticulocyte lysate (TNT systems, Promega) according to the manufacturer’s instruction. GST pull-down was performed using 5 µg of GST-USP11 with 8 µL of the transcribed/translated EZH2 and 30 µL of glutathione-Sepharose beads.
Western blot analysis
Protein extracts were lysed in SDS lysis buffer (50 mM Tris/HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% SDS), followed by centrifugation, the supernatants were collected. Equal amounts of protein (30 µg) in each group were resolved on 10% SDS-polyacrylamide gel electrophoresis. Separated proteins were transferred to polyvinylidene difluoride membranes followed by blocked with 5% non-fat milk and probed with antibodies against EZH2 (1:1,000), USP11 (1:500) or β-actin (1:2,000). Finally, the blots were incubated with peroxidase-conjugated secondary antibody against mouse or against rabbit (1:3,000). The immunoblots were visualized by the enhanced chemiluminescence assay (WBKLS0500, Millipore) and the intensity was determined using the Image-Pro Plus 6.0 software (Japan). β-actin was used as an internal normalization control.
Cell growth analysis
The breast cancer cell line MCF-7 was plated into 6-well culture plates (5×104 cells/well), and incubated in a humidified atmosphere of 5% CO2 at 37 °C. Cell growth rate was monitored at different time intervals by counting cell numbers. Independent experiments were carried out in triplicate.
Transwell invasion assay
The invasive ability of breast cancer cells was measured using Transwell assay (Chemicon Incorporation), and matrigel were from BD Biosciences. After transfected with the relative siRNA, 2×105 MDA-MB-231 cells in 100 µL serum free media were added onto the upper chamber. The lower compartment was incubated with RPMI 1640 containing 10% fetal bovine serum. MDA-MB-231 cells were incubated for 24 hours at 37 °C. The cells invaded into the lower surface were fixed with 2% paraformaldehyde, stained with 0.5% crystal violet and counted for five different fields under a microscope.
MRI detection
For in vivo MRI, MCF-7 siUSP7 cells, SCR cells, Vector cells and USP7 cells were used, and the details of the protocol were mentioned early. Mice were sacrificed and blood T1’s determined from samples were taken from the inferior vena cava (16).
Statistical analyses
All statistical analyses were done using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). P value <0.05 was considered as statistically significant. All results were expressed as mean ± SD, and were performed for at least three times independently. Student’s t-test was used to correlate two independent groups comparison. Kaplan-Meier survival curves were plotted to analyze the breast cancer morbidity. Univariate Cox regression analysis was used to assess the marginal effect. The differences between groups were assessed using log-rank analysis.
Results
The higher expression of USP11 indicated a poorer prognosis in breast cancer
In order to understand the role of USP11 in breast cancer, qRT-PCR was used to examine the USP11 mRNA expression level, as showed in Figure 1A, there was obviously increase in the USP11 expression in 120 breast cancer samples compared with normal breast tissues, (P<0.05). GAPDH was used as an internal control. The similar tendency was also observed in the breast cancer cell lines MCF-7 and MDA-MB-231 compared with the normal breast epithelial cell line MCF-10A, from both mRNA (Figure 1B) and protein level (Figure 1C). Depending on the median USP11 mRNA expression level, the breast cancer patients were divided in to two groups, and the higher expression of USP11 was with a poorer prognosis for overall 5-year survival times (P<0.01) (Figure 1D). As summarized in Table 1, the higher USP11 expression was significantly associated with higher histological grade, ER negative, higher clinical stage and Lymph node metastasis breast cancer, all of which indicated worse prognosis.
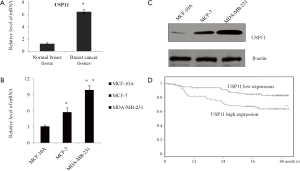
Full table
USP11 promoted breast cancer cells proliferation in vitro
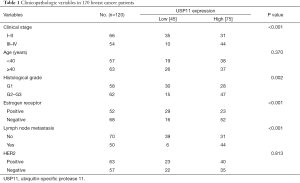
To explore the function of USP11 in breast cancer cells, firstly, MCF-7 cells as well as MDA-MB-231 cells were infected with control siRNA, siUSP11#1, or siUSP11#2, the knockdown efficiency was detected. Cells transfected with siUSP11 were significantly reduced in the USP11 mRNA level compared with the control groups (Figure 2A). Concomitantly, in the USP11 protein level, the siUSP11 groups were also markedly lower than the control groups as detected by western blot analysis (Figure 2B). siUSP11#2 was chosen for the further experiments because of better efficiency. USP11 over expression efficiency was also detected in MCF-7 cells or MDA-MB-231 cells transfected with vector or USP11 construct (Figure 2C). Cell growth assays was performed in MCF-7 cells transfected with vector, USP11 gain-of-function, USP11 loss-of-function. As showed in Figure 2D, MCF-7 cells with USP11 over expression showed an obviously growth promotion, while in the USP11 knockdown group, there was an evident growth inhibition, compared with the untreated group. Cell growth assays using MDA-MB-231 cell lines further proved the function of USP11 in promoting proliferation in vitro (Figure 2E).
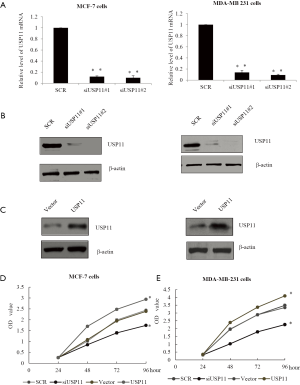
USP11 promoted breast cancer cells invasion and enhanced MRI detection
In order to explore the role of USP11 in the migration and invasion, transwell assay was conducted, as an approach to study invasion. MDA-MB-231 cells were pretreated with control siRNA or siUSP11#2, or vector, or USP11 plasmid. Cells transmigrated to the lower chamber were counted, as showed, after infection with siUSP11 #2, the migrated cells were significantly decreased. While after infection with USP11, the migrated cells were obviously increased. Altogether, the above experiments indicated USP11 caused a notable increase in migrating cells (Figure 3A,B). In the vivo MRI assay, as measured, in the USP11 knockdown group, the permeability-surface area product was hardly to detected (Figure 3C). Oppositely, USP11 over expression resulted in the noninvasive MCF-7 to a more detectable phenotype, as the permeability-surface area product was significantly higher compared with the vector (Figure 3D).
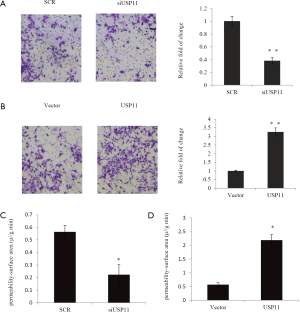
EZH2 was identified as a USP11-interacting protein
Since USP11 was a deubiquitylating enzyme, we tried to investigate the potential substrates of USP11 in breast cancer. Co-immunoprecipitation (co-IP) assay was performed to identify the USP11-interacting proteins in MCF-7 cells. Several key regulators that have been reported to work as breast cancer driver were chosen for study such as transcription factors and histone modification enzymes. As showed in Figure 4A, JenyH3 lysine 27 (H3K27) trimethyltransferase EZH2 could interact with USP11 in vivo. Since firstly immune precipitated with antibodies against USP11, and then followed by western blotting against EZH2 antibody, there was obviously band compared with the normal IgG group. Reciprocal immunoprecipitation with EZH2 antibody and immunoblotting with antibody against USP11 also indicated the fact. The interaction was also observed in MDA-MB-231 cells with endogenous proteins (Figure 4B). The above data indicated that USP11 interacted with EZH2 in vivo. Further, GST pull-down assay was performed to investigate whether USP11 could interact with EZH2 directly. Glutathione S-transferase fusion protein USP11 was immobilized on glutathione Sepharose 4B beads, and incubated with translated FLAG-tagged EZH2, with GST protein used as a NC. As showed in Figure 4C, EZH2 was found to bind with USP11 directly in vitro. Collectively, the above experiments indicated that USP11 was physically associated with EZH2 in vivo and in vitro.

USP11 inhibited EZH2 ubiquitination and stabilized EZH2 in vivo
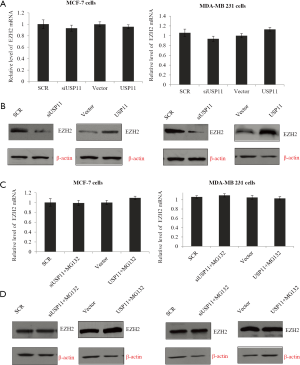
Since USP11 was reported to deubiquitinate and stabilize BRCA2 and ALK5, it was reasonable to make the hypothesis that USP11 might also regulate EZH2 via its deubiquitinase activity. To explore this, MCF-7 cells or MDA-MB-231 cells were infected with control siRNA, siUSP11#2, vector, or USP11 separately, and the expression of EZH2 was detected. As expected, although the mRNA expression was not changed (Figure 5A), in the USP11 knockdown group, the protein level of EZH2 decreased notably, while in the USP11 over expression group, EZH2 expression increased remarkably (Figure 5B). Those indicated USP11 may influence EZH2 through post transcription modification. To determine whether USP11 could influence EZH2 expression through protein ubiquitination pathway, MG132, a proteasome-specific inhibitor, was used. In the USP11 knockdown MCF-7 or MDA-MB-231 cells, MG132 was successful to rescue EZH2 protein from degradation (Figure 5C,D). Based on the above experiments, we made the conclusion that USP11 stabilized the expression of EZH2 possibly based on its deubiquitinating activity.
Discussion
Several groups have reported that USP11 could stabilize the target genes by deubiquitinating the polyubiquitinated substrates, for example, USP11 regulated p53 stability by deubiquitinating p53 (17). Similarly, USP11 negatively regulated NF-kappaB activation by catalyzing deubiquitination of IkappaBalpha (18). So we made the hypothesis that more and more substrates regulated by USP11 will be identified in the future.
Although lower expression of USP11 was correlated with better survival outcomes in breast cancer (15), the mechanism was still unclear. More evidence needs to be excavated to understand the role of USP11 in breast cancer. In the present study, we first investigated the expression of USP11 in 120 cases of breast cancer and the adjacent normal tissues, our data demonstrated that USP11 mRNA expression was higher in the cancer samples compared with the normal tissues. We defined the function of USP11 in the breast cancer cell proliferation and invasion. Using MCF-7 and MDA-MB-231 cell lines, the ectopic expression of USP11 could increase the proliferation ability of MCF-7 cells, the invasion ability of MDA-MB-231 cells and enhance the MRI detection, while the knockdown of USP11 opposite the tendency. One of the mechanisms was because USP11 could interact with EZH2 in vivo and in vitro directly. Another evidence to support our argument was that over expression of USP11 could result in the enrichment in the protein level of EZH2 but not the mRNA level, which further indicated that regulation of EZH2 by USP11 was due to post-transcriptional modification, possibly based on its deubiquitinating activity. As it has been well studied that EZH2 worked as a breast cancer tumorigenesis driver (19-21), it was reasonable that USP11 might play an important role in breast cancer progression and work as a new therapeutic target in triggering breast cancer by stabilization of EZH2.
Acknowledgments
Funding: This work was supported by a Grant for the Science and Technology Project of Suzhou (SYS201614).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.01.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written consent was obtained from all participants prior to the study. The ethical approval was given by the hospital ethics committee and the ethics approval ID was Suzhou University-Ethics-D-20150831.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol 2011;223:307-17. [Crossref] [PubMed]
- Hong W, Dong E. The past, present and future of breast cancer research in China. Cancer Lett 2014;351:1-5. [Crossref] [PubMed]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002;298:1039-43. [Crossref] [PubMed]
- Asangani IA, Ateeq B, Cao Q, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell 2013;49:80-93. [Crossref] [PubMed]
- Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012;338:1465-9. [Crossref] [PubMed]
- Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol 2006;24:268-73. [Crossref] [PubMed]
- Chang CJ, Yang JY, Xia W, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell 2011;19:86-100. [Crossref] [PubMed]
- Yu H, Simons DL, Segall I, et al. PRC2/EED-EZH2 complex is up-regulated in breast cancer lymph node metastasis compared to primary tumor and correlates with tumor proliferation in situ. PLoS One 2012;7:e51239 [Crossref] [PubMed]
- Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer 2012;106:243-7. [Crossref] [PubMed]
- Chen S, Huang L, Sun K, et al. Enhancer of zeste homolog 2 as an independent prognostic marker for cancer: a meta-analysis. PLoS One 2015;10:e0125480 [Crossref] [PubMed]
- Lin CH, Chang HS, Yu WC. USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. J Biol Chem 2008;283:15681-8. [Crossref] [PubMed]
- Schoenfeld AR, Apgar S, Dolios G, et al. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol 2004;24:7444-55. [Crossref] [PubMed]
- Al-Salihi MA, Herhaus L, Macartney T, et al. USP11 augments TGFβ signalling by deubiquitylating ALK5. Open Biol 2012;2:120063 [Crossref] [PubMed]
- Bayraktar S, Gutierrez Barrera AM, Liu D, et al. USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer J 2013;19:10-7. [Crossref] [PubMed]
- Pathak AP, McNutt S, Shah T, et al. In vivo "MRI phenotyping" reveals changes in extracellular matrix transport and vascularization that mediate VEGF-driven increase in breast cancer metastasis. PLoS One 2013;8:e63146 [Crossref] [PubMed]
- Ke JY, Dai CJ, Wu WL, et al. USP11 regulates p53 stability by deubiquitinating p53. J Zhejiang Univ Sci B 2014;15:1032-8. [Crossref] [PubMed]
- Sun W, Tan X, Shi Y, et al. USP11 negatively regulates TNFalpha-induced NF-kappaB activation by targeting on IkappaBalpha. Cell Signal 2010;22:386-94. [Crossref] [PubMed]
- Gonzalez ME, Moore HM, Li X, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A 2014;111:3098-103. [Crossref] [PubMed]
- Mu Z, Li H, Fernandez SV, et al. EZH2 knockdown suppresses the growth and invasion of human inflammatory breast cancer cells. J Exp Clin Cancer Res 2013;32:70. [Crossref] [PubMed]
- Moore HM, Gonzalez ME, Toy KA, et al. EZH2 inhibition decreases p38 signaling and suppresses breast cancer motility and metastasis. Breast Cancer Res Treat 2013;138:741-52. [Crossref] [PubMed]


