Gene signatures predictive of response to radiotherapy in prostate cancer: a new step towards precision medicine
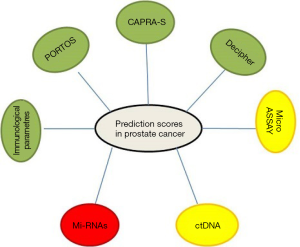
In the era of personalized medicine, there is significant emphasis on the development of companion diagnostics and/or molecular signatures to guide therapeutic decisions (1). For example, two recurrence risk signatures (Oncotype Dx® and Mammaprint) are commonly used to guide chemotherapy in women with node-negative breast cancer (2-5). An evolution of Oncotype® (that is a centralized method of determination of the gene signature in breast cancer) is EndoPredict. The latter is a not centralized method of genotyping of the breast cancer that associates a signature of genes to the clinical staging of the patients and that was recently demonstrated to give higher performance if compared with the other well established genotyping system Oncotype. In fact, the test can predict whether breast cancer will spread in women with estrogen receptor positive, HER2 negative (ER+/HER2–) disease (6). Women with ER+/HER2– breast cancer are given endocrine therapy after surgery to treat their cancer. They receive also chemotherapy if there is a reasonable risk of the cancer spreading to other organs (referred to as secondary or metastatic breast cancer). This is because if the cancer spreads to certain organs, it may not be possible to treat it and the cancer can become incurable. EndoPredict analyses the activity of eight different genes within a tumor sample, and uses this information alongside the patient’s tumor size and nodal status to give an ‘EPclin’ score estimating their risk of developing advanced breast cancer. The EPclin score is then used to categorize patients into low or high risk groups, with a cut-off point of 10% risk over 10 years. When Oncotype DX was used to identify the third of lymph node-negative patients with the lowest risk of secondary disease, 7% went on to develop secondary breast cancer after 10 years. Of the third of patients identified as having the lowest risk according to the EPclin score, only one of 227 (0.5%) patients developed secondary disease. By identifying a large group of patients with a very good prognosis, EPclin could offer to clinicians and particularly to patients a reliable reassurance that chemotherapy can be avoided. In the field of the prediction of response to anti-tumor agents, K-ras and N-Ras mutations has been shown to be predictive of panitumimab and cetuximab nonbenefit in colorectal cancer (7,8). Furthermore, epidermal growth factor receptor (EGFR) mutations have been shown to predict benefit from tyrosine kinase inhibitors (TKI) and more recently, ALK gene rearrangement has shown to be predictive for crizotinib benefit in non-small cell lung cancer (NSCLC) (9-11). Moreover, fibroblast growth factor receptor (FGFR) mutations and/or amplification are emerging as new predictor markers of response to agents raised against FGFR in a variety of tumors, e.g., adeno- and squamous cell carcinoma of the lung, glioblastoma multiforme and bladder cancer. FGFR3 mutations are reported in up to 50% of cancers of all stages from the lower and upper urinary tract with p.S249C being the most common mutation, found in 61% of cases. Mutation is inversely correlated with tumor stage and grade, and mutated tumors are associated with a favourable clinical outcome. The mutation analysis potentially allows a further stratification of patients, and should be additional evaluated in larger cohorts of invasive tumors. Bearing in mind that recurrence-free survival is an indicator for disease severity and risk of progression, FGFR3 mutations in squamous differentiated bladder tumors may indicate potential for FGFR inhibitor treatment in these tumors (12). On the other hand, clinical decision making in radiation oncology is still mainly based only on clinicopathologic features. Therefore, there is a great need to develop molecular diagnostics to more efficiently use radiotherapy (RT). Effective predictive biomarkers are a central requirement for the development of personalized treatment in clinical oncology. Unlike prognostic biomarkers, which predict clinical outcome independently from treatment, predictive biomarkers are treatment specific and thus are critical for therapeutic decision making (13). For example, several targeted drugs are now routinely offered to patients whose tumors harbour a specific marker for benefit or nonbenefit [i.e., HER-2/neu expression and trastuzumab benefit (14), K-ras mutation, and panitumumab nonbenefit] (7). In contrast, radiation therapy is still recommended on the basis of standard clinicopathologic features, which generally address tumor burden/aggressiveness and serve as prognostic biomarkers of outcome rather than a specific marker for RT therapeutic benefit. It is estimated that approximately a third of patients with localized or locally advanced prostate cancer undergo external beam RT with curative intent (15). The use of RT in combination with androgen-deprivation prolongs survival (16), and has contributed to the increase in 5-year survival rate from 30% in the 1970s to 80% in 2009 (17). Late toxicity following irradiation for prostate cancer includes damage to the bladder, bowel and erectile function. The median rates of late gastrointestinal (GI) and genitourinary (GU) toxicity are reported to be 15% and 17%, respectively (18). Studies are attempting to identify the genetic variants that increase an individual’s risk of radiation toxicity (19,20). Moreover, a low number of studies have made efforts to identify biological features of prostate cancer tissues and immunologic circulating biomarkers able to identify patients responsive to RT. In this light, Nardone et al. (21) have recently found that tumor infiltration by different lymphocyte subsets predicts the outcome of patients with prostate cancer showing only local relapse after primary surgery and subsequently receiving RT. They establish a premise for a possible immunological therapy associated with RT for selected patients. In fact, the chemotherapy/RT could induce DNA double-strand breaks that, in turn, produce mutations and neo-antigens generation, promote immunological danger signals and reduce tumor infiltrating immunosuppressive cell populations, such as inhibitory myeloid cells. All these events are necessary to trigger antigen-specific CTLs. The critical role of the tumor immunologic microenvironment in conditioning both tumor development and survival offers the rationale to design new immunotherapeutic strategies for patients with prostate cancer associated to radiation treatment. On the light of the genetic scores of prostate cancer, in a matched retrospective analysis reported in The Lancet Oncology, Zhao et al. (22) identified and validated a 24-gene predictor of response to postoperative RT in prostate cancer. In the training cohort (n=196 from one study) and pooled validation cohort (n=330 from four remaining studies), patients who had post-operative RT were matched with patients who did not receive RT based upon clinical and pathological parameters including Gleason score, PSA level, surgical margin status, extracapsular extension, seminal vesicle invasion, lymph node invasion, and androgen-deprivation therapy. In the training cohort, a 24-gene Post-Operative Radiation Therapy Outcomes Score (PORTOS) was used to predict response to post-operative RT. The 24-gene set included 6 genes related only to DNA-damage response, 4 genes related to both DNA-damage and radiation response, and several genes involved in immune response (including IL1B, IL7R, PTPN22, and HCLS1). The primary endpoint was development of distant metastasis. They used high-throughput gene expression and clinical data to develop and validate 24-gene expression signature that predicts response to post-prostatectomy RT (PORTOS) in matched training and validation cohorts of patients with prostate cancer. They show that, in patients receiving RT, patients with high levels of 24-gene expression had a lower incidence of distant metastasis than in patients with low scores. In the same study, the new signature score PORTOS was compared to the predictive value of the already standardized methods of prediction Decipher, CAPRA-S, and microarray version of the cell cycle progression (CCP) signature. Decipher is a genomic test, which evaluates the activity of genes in the tumor that are shown to be involved in the development and progression of prostate cancer. In details, Decipher measures the expression levels of 22 RNA biomarkers involved in multiple biological pathways across the genome that is associated with aggressive prostate cancer. CAPRA-S is a straightforward instrument for facilitating disease risk classification. A CAPRA score is valid across multiple treatment approaches and predicts an individual’s likelihood of metastasis, cancer-specific mortality, and overall mortality. The score is calculated using points assigned to: age at diagnosis, PSA at diagnosis, Gleason score of the biopsy, clinical stage and percent of biopsy cores involved with cancer (again clinical and pathological features of the tumor). Genes whose expression is regulated as a function of CCP were originally identified as having RNA expression levels that oscillated as cells progressed through various stages of the cell cycle. Since the expression levels of CCP genes probably reflect fundamental aspects of tumor biology, 31 CCP genes were selected and tested for their ability to predict disease outcome using a predefined score based on their expression levels. Using the median score as the cutoff point, the interactions between the Decipher, mCCP, and CAPRA-S prognostic models with RT were not significant (interaction between RT and score, Decipher P interaction =0.99, mCCP P interaction =0.34, CAPRA-S P interaction =0.34). In details, patients with high Decipher, mCCP, or CAPRA-S scores do worse than do those with a low score regardless of treatment, and patients treated with RT have improved outcomes regardless of risk score. In conclusion in this interesting report, in comparison to PORTOS, the widely used genomic and clinical risk tools Decipher, mCCP, and CAPRA-S did not predict response to post-operative RT. However, a combination of Decipher and PORTOS could allow for selection of patients who need post-operative RT (using PORTOS), and help decide whether to irradiate in the adjuvant or salvage setting (using Decipher). These evaluations were not conducted in routine clinical settings. No evidence was identified to address the question of clinical utility. Future research should focus on evaluating clinical validity more extensively and robustly in the general clinical populations, and on comparing PORTOS panel directly with the existing standard care and diagnostic standards. In addition, emphasis should be given to the finding of new circulating genetic biomarkers that can be easily assessed through not invasive procedures in prostate cancer patients. In these lights, circulating micro-RNAs and circulating tumor DNA (ctDNA) were recently investigated. The following circulating miRNAs were found to be associated to the development and progression of prostate cancer. miR-21 expression increases together with clinical parameters (such as Gleason score or lymph node metastases) and is correlated with castration resistance and metastatic disease. MiR-21 and miR-18 are also useful as biomarkers in prediction of progression of prostate cancer. Another oncogenic miRNA overexpressed in prostate cancer and positively correlated with poor overall and PSA recurrence free survival, is miR-4534. It is hypermethylated in normal cells and tissues compared to those of prostate cancer and exert its oncogenic effects partly by downregulating the tumor suppressor PTEN gene. Its overexpression induces pro-cancerous characteristics in non-cancer cell line whereas its knockdown impair cell proliferation, migration/invasion and induce G0/G1 cell cycle arrest and apoptosis in prostate cancer. MiR-32 is highly expressed in castration resistant prostate cancer (CRPC) samples compared to benign prostatic hyperplasia samples. The reduction of miR-145 expression in prostate cancer was correlated with higher Gleason scores, advanced clinical stage, larger tumor diameter and higher PSA and follow-up PSA levels. miRNAs are important modulators of gene expression. They are frequently altered in prostate cancer and as such offer the potential to be used as biomarkers or novel therapeutic targets. However, none of them are still validated for clinical use. Regarding the cell-free ctDNA and circulating tumor cells (CTCs) are plasma sources of tumor DNA that have been investigated for non-invasive detection and monitoring of patient tumors but have not been analyzed or directly compared across multiple tumor types. ctDNA liquid biopsy allows to understand specifically what kind of molecular changes are happening in the tumor in real time, which is a very big step beyond where CTCs are today in clinical terms. Perhaps the most promising applications of CTCs and ctDNA are molecular analyses that can inform the rational selection of appropriate therapies for patients. In example, in the treatment of a patient with prostate cancer, alterations in the androgen receptor variant 7 mutation, detected in CTCs, predicts the lack of response to abiraterone or enzalutamide and can be useful to provide the most immediately actionable information regarding the choice between AR-targeted therapies or non-AR-targeted therapies such as cytotoxic chemotherapy (for a summary of the scores available for the prediction of response in prostate cancer see Figure 1) (23). The use of this information in real time can guide the clinicians in the choice of the best-personalized therapy in the case of androgen depletion therapy. It has also to be considered that PORTOS diagnostic system can open a new scenario of investigations coming back to the bench in order to study if any of the evaluated genes can be efficiently assessed directly in the blood of the patients and if some mutations can be revealed that correlate to the response to post-operative RT, for which the information is still limited. Another chance given by the authors of the manuscript by Zhao et al. is to integrate PORTOS score with other circulating miRNAs and/or ctDNA and/or CTCs in order to increase the prognostic accuracy in the same set of patients.
In conclusion, the manuscript by Zhao et al. disclosed the possibility to study genetic scores of the prostate cancer that correlates to the response to post-operative RT independently from the conventional clinical and pathological features of the disease and strongly encourages additional studies on new intratumor or circulating biomarkers in this subset of patients.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.02.25). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet 2009;10:77-82. [Crossref] [PubMed]
- Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin 2011;61:91-112. [Crossref] [PubMed]
- van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999-2009. [Crossref] [PubMed]
- van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530-6. [Crossref] [PubMed]
- Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817-26. [Crossref] [PubMed]
- Buus R, Sestak I, Kronenwett R, et al. Comparison of EndoPredict and EPclin With Oncotype DX Recurrence Score for Prediction of Risk of Distant Recurrence After Endocrine Therapy. J Natl Cancer Inst 2016;108:djw149 [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011;12:1004-12. [Crossref] [PubMed]
- Baldia PH, Maurer A, Heide T, et al. Fibroblast growth factor receptor (FGFR) alterations in squamous differentiated bladder cancer: a putative therapeutic target for a small subgroup. Oncotarget 2016;7:71429-39. [PubMed]
- Clark GM. Prognostic factors versus predictive factors: Examples from a clinical trial of erlotinib. Mol Oncol 2008;1:406-12. [Crossref] [PubMed]
- Baselga J. Treatment of HER2-overexpressing breast cancer. Ann Oncol 2010;21:vii36-40. [Crossref] [PubMed]
- Foroudi F, Tyldesley S, Barbera L, et al. Evidence-based estimate of appropriate radiotherapy utilization rate for prostate cancer. Int J Radiat Oncol Biol Phys 2003;55:51-63. [Crossref] [PubMed]
- Martin NE, D'Amico AV. Progress and controversies: Radiation therapy for prostate cancer. CA Cancer J Clin 2014;64:389-407. [Crossref] [PubMed]
- Cancer Research UK Prostate Cancer Key Stats. Available online: http://www.cancerresearchuk.org/cancer-info/cancerstats/keyfacts/ prostate-cancer/, accessed February 2015.
- Ohri N, Dicker AP, Showalter TN. Late toxicity rates following definitive radiotherapy for prostate cancer. Can J Urol 2012;19:6373-80. [PubMed]
- Wilkins A, Mossop H, Syndikus I, et al. Hypofractionated radiotherapy versus conventionally fractionated radiotherapy for patients with intermediate-risk localised prostate cancer: 2-year patient-reported outcomes of the randomised, non-inferiority, phase 3 CHHiP trial. Lancet Oncol 2015;16:1605-16. [Crossref] [PubMed]
- Dolezel M, Odrazka K, Zouhar M, et al. Comparing morbidity and cancer control after 3D-conformal (70/74 Gy) and intensity modulated radiotherapy (78/82 Gy) for prostate cancer. Strahlenther Onkol 2015;191:338-46. [Crossref] [PubMed]
- Nardone V, Botta C, Caraglia M, et al. Tumor infiltrating T lymphocytes expressing FoxP3, CCR7 or PD-1 predict the outcome of prostate cancer patients subjected to salvage radiotherapy after biochemical relapse. Cancer Biol Ther 2016;17:1213-20. [Crossref] [PubMed]
- Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol 2016;17:1612-20. [Crossref] [PubMed]
- Miyamoto DT, Lee RJ. Cell-free and circulating tumor cell-based biomarkers in men with metastatic prostate cancer: Tools for real-time precision medicine? Urol Oncol 2016;34:490-501. [Crossref] [PubMed]