
DNA ligases as therapeutic targets
Introduction
The identification of DNA repair defects in inherited human diseases that are characterized by predisposition to cancer, including inherited forms of colon and breast cancer, provides compelling evidence that the cellular mechanisms that maintain genome stability play a critical role in suppressing cancer formation (1). Since genomic instability is a hallmark feature of sporadic as well as hereditary cancers, it is likely that alterations in one or more of the mechanisms that maintain genome stability occur at some stage during the development of most cancers. Although it has been assumed that these alterations in the DNA damage response contribute, at least in part, to the therapeutic activity of cytotoxic DNA damaging agents such as cis-platinum and doxorubicin, they remain poorly characterized, particularly in sporadic cancers. The recent development of poly (ADP-ribose) polymerase inhibitors as therapeutics that selectively target the DNA repair defect in hereditary breast cancers has stimulated interest in defining abnormalities in the DNA damage response in sporadic cancers (2) and the development of inhibitors of other DNA repair proteins that may have utility as anti-cancer agents (2). Since DNA joining is required to complete almost all DNA repair events and there are three human genes encoding DNA ligases with different but overlapping functions in DNA replication and repair (3), DNA ligase inhibitors with defined specificity can potentially be combined with different DNA damaging agents to target a wide variety of DNA repair pathways. In this review, we summarize our current understanding of the cellular functions of human DNA ligases (Table 1) and recent studies that identify DNA ligases as potential biomarkers for abnormal DNA repair and demonstrate the potential clinical utility of DNA ligase inhibitors in cancer treatment.
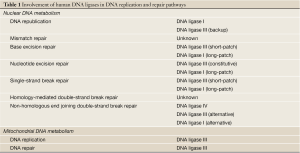
Full table
Structure and function of human DNA ligases
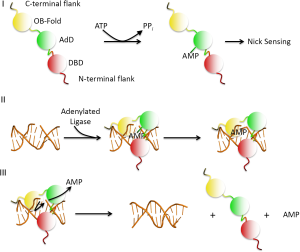
DNA ligases maintain the integrity of the phosphodiester backbone of duplex DNA by catalyzing phosphodiester bond formation (3). All DNA ligases utilize the same three-step reaction mechanism (Figure 1). In humans, the DNA ligases encoded by the three LIG genes are ATP-dependent (3). In step 1, ATP is hydrolyzed, resulting in the covalent linkage of an AMP moiety to a specific lysine residue within the DNA ligase active site and the release of pyrophosphate. Next, the AMP moiety is transferred from the adenylated ligase to the 5' terminus of a DNA nick with 5' phosphate and 3' hydroxyl termini, generating a DNA-adenylate intermediate. Finally, the non-adenylated DNA ligase interacts with the DNA adenylate and, using the 3' hydroxyl as a nucleophile, links the termini via a phosphodiester bond, releasing AMP.
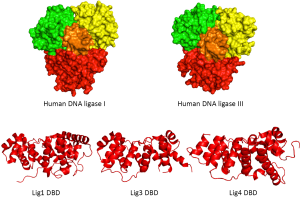
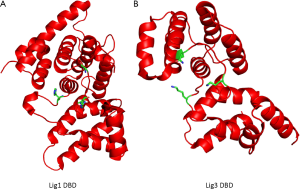
Human DNA ligases contain a common catalytic core consisting of an oligonucleotide/oligosaccharide binding-fold (OB-fold) domain and an adenylation domain (AdD) that are found in all DNA ligases and other nucleotidyl transferases including RNA ligases and mRNA capping enzymes (3). While these two domains comprise the minimum unit that can perform the DNA ligation reaction, the activity of the catalytic core of human DNA ligases is greatly enhanced by an additional conserved N-terminal DNA-binding domain (DBD) (3,4). In the absence of DNA, the catalytic region of human DNA ligases encompassing the OB-fold, AdD and DBD adopts an extended, asymmetric conformation (3,5). As shown in Figure 2A, the structures of the catalytic regions of DNA ligase I and DNA ligase III bound to non-ligatable nicked DNA, which were determined by X-ray crystallography (4,6), revealed that these enzymes form similar ring-shaped structures around nicked DNA. Thus, the catalytic regions of the DNA ligases are flexible and undergo large conformational changes during the ligation reaction (Figure 1). It is assumed that the catalytic region of DNA ligase IV will behave in a similar manner. Recently, the structure of the DBD of DNA ligase IV was determined by X-ray crystallography in the absence of DNA (7). As expected, the DBD of DNA ligase IV has a similar overall structure to the DBDs of DNA ligases I and III (Figure 2B). Notably, the availability of atomic resolution structural information has permitted the use of rational structure-based approaches to identify small molecule inhibitors of human DNA ligases. The properties of these inhibitors and their potential as cancer therapeutics are discussed later in this article.
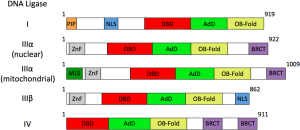
In contrast to the DBD and catalytic core of human DNA ligases I, III and IV, the regions in these enzymes adjacent to the catalytic region are much more diverse. Furthermore, unlike the LIG1 and LIG4 genes, the LIG3 gene encodes multiple DNA ligase polypeptides that have different N- and C-terminal regions (Figure 3). An alternative translation initiation mechanism generates polypeptides that either have or lack an N-terminal mitochondrial localization signal from LIG3 mRNA (8). This signal sequence is removed during entry into mitochondria and so the nuclear and mitochondrial versions of DNA ligase III encoded by this mRNA transcript are very similar in size. In addition, a germ cell-specific alternative splicing mechanism generates polypeptides with different C-terminal sequences (9). At the C-terminus of the DNA ligase IIIα polypeptide there is a breast and ovarian cancer susceptibility protein 1 C-terminal (BRCT) domain that mediates an interaction with the C-terminal BRCT domain of nuclear DNA repair protein X-ray cross complementing protein 1 (XRCC1) (9-12). Structural analysis of the BRCT-BRCT heterodimer revealed that residues adjacent to the XRCC1 domain contribute to heterodimer interface, favoring formation of the DNA ligase IIIα/XRCC1 heterodimers rather than homodimers of DNA ligase IIIα and XRCC1 (13). In the DNA ligase IIIβ polypeptide, the C-terminal BRCT domain is replaced by a short sequence that acts as a nuclear localization signal (9). All the DNA ligase III polypeptides have an N-terminal zinc finger domain (ZnF) that, in concert with the DBD, plays a key role both in the initial recognition of DNA strand breaks and intermolecular ligation (14-16). BRCT domains also occur in DNA ligase IV. In this case, there are two tandemly arrayed BRCT domains within the C-terminal region of DNA ligase IV. As with DNA ligase IIIα, the DNA ligase IV BRCT domains are critical for an interaction with a partner protein, the DNA repair protein X-ray cross complementing protein 4 (XRCC4) (17-19). Structural studies have shown that within the DNA ligase IV/XRCC4 complex, the two BRCT domains encircle the coiled-coil region of the XRCC4 homodimer and that the linker region between the BRCT domains of DNA ligase IV makes the majority of the contacts with XRCC4 (20-22). The only recognizable motifs within the N-terminal region of DNA ligase I are a nuclear localization signal and a proliferating cell nuclear antigen (PCNA) interacting protein (PIP) box which, as the name suggests, interacts with PCNA (23-25).
Cellular functions of human DNA ligases
Nuclear DNA replication
There is compelling evidence that DNA ligase I is the replicative DNA ligase (3,26). Human DNA ligase I is recruited to nuclear DNA replication foci via its PIP box-dependent interaction with PCNA (24). In addition, it also interacts with replication factor C (RFC), a clamp loader that loads the trimeric PCNA ring onto DNA during Okazaki fragment synthesis (27,28). Finally, cell lines (46BR and 46BR.1G1) established from an immunodeficient human patient with mutated LIG1 alleles have reduced DNA ligase I activity and severely impaired Okazaki fragment joining (28-30). As expected, DNA ligase I activity is essential for embryogenesis in mice but, surprisingly, it was possible to establish cell lines from LIG1 null embryos that had a defect in joining Okazaki fragments similar to the human 46BR cell lines (31,32). The viability of LIG1 null cells indicates that another DNA ligase participates in nuclear DNA replication in the absence of DNA ligase I. Recent studies in chicken DT40 and human cells have shown that DNA ligase IIIα but not DNA ligase IV, is essential for nuclear DNA replication in the absence of DNA ligase I (33,34). XRCC1, which interacts with and stabilizes nuclear DNA ligase IIIα (10,12,35), and poly (ADP-ribose) polymerase 1 (PARP-1), which initiates the repair of DNA single strand breaks (SSB)s (36), are also required for nuclear DNA replication in DNA ligase I-deficient cells (33). Thus, it appears that, in the absence of DNA ligase I, Okazaki fragment joining is accomplished, at least in part, through a SSB repair mechanism in which ADP-ribosylated PARP-1 recruits the DNA ligase IIIα/XRCC1 complex via its interaction with XRCC1 (33).
Mitochondrial DNA replication and repair
Genetic inactivation of the mouse LIG3 gene result in embryonic lethality at an even earlier stage than LIG1 and XRCC1 null embryos (31,32,37,38). Furthermore, although LIG1 and XRCC1 mouse embryonic cell lines were established from the embryos (31,32,37), it was not possible to establish a LIG3 null cell line, suggesting that the LIG3 gene is required for cell viability (38). This has been confirmed in recent studies showing that the LIG3 gene is indeed essential because it encodes the only mitochondrial DNA ligase (39,40). These observations were predicted by earlier studies showing that alternative translation initiation generates nuclear and mitochondrial versions of DNA ligase IIIα and that mitochondrial DNA ligase IIIα functions independently of XRCC1 (8,41). Furthermore, depletion of DNA ligase IIIα levels by siRNA resulted in reduced numbers of copies of the mitochondrial genome and increased accumulation of SSBs in mitochondrial DNA (42). Thus, mitochondrial DNA ligase IIIα plays an essential and unique role in the replication and repair of the mitochondrial genome (Table 1).
Nuclear DNA excision repair
There are three pathways that excise mismatched and/or damaged bases from DNA: mismatch repair (MMR), base excision repair (BER) and nucleotide excision repair (NER). The major function of the MMR pathway is to correct mistakes in newly synthesized DNA made by the DNA replication machinery (1). Although significant progress has been made in elucidating the molecular mechanism of MMR, the identity of the DNA ligase (or DNA ligases) that completes this repair pathway has not been established definitively.
There is compelling evidence linking both DNA ligase I and DNA ligase IIIα with BER. This repair pathway is initiated when a DNA glycosylase recognizes and excises a damaged base (1). After the resultant abasic site is cleaved, there are two possible subpathways to complete the repair (3,43-45). In short-patch BER, DNA polymerase β removes the remaining 5' sugar-phosphate residue and inserts a single nucleotide, generating a ligatable nick that is sealed by the DNA ligase IIIα/XRCC1 complex (3,44). This pathway is thought to be active across the entire genome in non-dividing cells and throughout the cell cycle. In contrast, the long-path BER pathway, in which a longer repair patch is inserted by either DNA polymerase δ or ε and repair is completed by the action of DNA ligase I in conjunction with FEN-1, appears to occur only during S phase and to be linked to the DNA replication machinery (3,43,45). Analysis of DNA ligase-deficient human and mouse cells have given paradoxical results regarding the contribution of DNA ligase I- and DNA ligase IIIα-dependent BER to cell survival after DNA alkylation damage. For example, human DNA ligase I-deficient 46BR cells are sensitive to DNA alkylating agents (28,46,47) whereas LIG1 null mouse embryonic fibroblasts and mouse embryonic fibroblasts that express a mutant version of DNA ligase I that is equivalent to the enzyme expressed in 46BR cells are not (31,32,48). This could be explained if DNA ligase I-dependent BER predominates in human cells whereas DNA ligase IIIα-dependent BER predominates in mouse cells. However, the characterization of mouse cells lacking nuclear DNA ligase IIIα led to the conclusion that DNA ligase I-dependent BER is the major pathway in the mouse cell-types examined (39,40). These discrepancies may reflect differences in the relative contributions of DNA ligase I- and DNA ligase IIIα-dependent BER and the extent of the functional redundancy between these BER subpathways in different cell types.
Nucleotide excision repair (NER) removes helix-distorting lesions such as ultraviolet light (UV)-induced photoproducts (1). In contrast to BER, the DNA lesion is removed by the excision of an oligonucleotide 24-32 nucleotides in length, followed by repair synthesis and ligation (1). For many years it was assumed that human DNA ligase I completed NER ligation because of the UV light sensitivity of 46BR cells (46) and the activity of DNA ligase I in conjunction with the replicative DNA polymerases δ and ε in reconstituted NER reactions (49,50). However, more recent studies have shown that DNA ligase I only accumulates at NER sites in proliferating cells, whereas DNA ligase IIIα and XRCC1 are recruited to NER sites regardless of cell cycle stage (51). Thus, DNA ligase I appears to function in a S phase-specific subpathway of NER, whereas the DNA ligase IIIα/XRCC1 complex functions in an NER subpathway that is active in all phases of the cell cycle and presumably in non-dividing cells (51). As with BER, the relative contributions of DNA ligase I- and DNA ligase IIIα-dependent NER and the extent of the functional redundancy between these NER subpathways may vary between different cell types.
Nuclear SSB repair
SSBs are generated in numerous different ways, including as repair intermediates during excision repair, by erroneous topoisomerase I activity, and by DNA damaging agents such as reactive oxygen species (ROS) and alkylating agents (52). As mentioned above, DNA SSBs are predominantly detected by PARP-1 although PARP-2 may also contribute (53,54). The binding of PARP-1 to SSBs activates its polymerase activity resulting in the synthesis of poly (ADP-ribose) chains on PARP-1 itself and other nearby proteins (1). The DNA ligase IIIα/XRCC1 complex is then recruited to SSBs primarily by an interaction between XRCC1 and poly (ADP-ribosylated) PARP-1 (36,55). Given the recruitment of XRCC1 and DNA ligase IIIα to SSBs, it was surprising that cells lacking nuclear DNA ligase IIIα did not exhibit a defect in SSB repair similar to cells with reduced levels of XRCC1 (39,40,56). Thus, it appears that there is an as yet poorly defined DNA ligase I-dependent SSB repair pathway (56).
Nuclear DSB repair
As with SSBs, DSBs are produced by many different mechanisms. For example, they arise during programmed cell events including meiotic recombination and immunoglobulin gene arrangements (1). They are also generated during normal DNA replication and, to an even greater extent, during replicative stress. Finally, they can be generated either directly by the action of a DNA damaging agent or indirectly as a consequence of the replication fork encountering an unrepaired SSB (1). There are multiple repair pathways for these highly cytotoxic lesions that can be divided into two groups depending upon whether or not the repair reaction involves extensive DNA sequence homology (57,58). While the DNA ligases that participate in the homology-dependent DSB repair pathways have not been definitively identified, it is well-established that DNA ligase IV is a key component of the major non-homologous end-joining (NHEJ) pathway, which is functional throughout the cell cycle, repairs most DSB lesions and completes V(D)J recombination (18). This pathway is initiated by the Ku70/Ku80 complex, which binds to DNA DSB ends and recruits the other components of the repair pathway, including DNA-dependent protein kinase catalytic subunit (DNA PKcs), Artemis and DNA ligase IV/XRCC4 (57). A key step in NHEJ is the juxtaposition of DNA ends that is mediated by interactions between DNA PKcs molecules (59). In addition, there are multiple end processing activities that act on the juxtaposed DNA ends to generate ligatable termini (57). As a consequence of this end porcessing, NHEJ is characterized by small insertions and deletions at the break site but usually the previously linked DNA ends are joined back together correctly (57).
Several DNA ligase IV-deficient individuals have been identified with symptoms that include radiation sensitivity, immunodeficiency and developmental delay (60-62). As with the LIG1 and LIG3 genes, genetic inactivation of LIG4 resulted in embryonic lethality in the mouse (63,64). Cells that lack LIG4 are viable (18,57), demonstrating that this repair pathway is not essential. Analysis of NHEJ-deficient cells revealed the presence of an alternative (alt) NHEJ pathway that also appears to be active in wild type cells albeit at a low level (65-68). Repair of DSBs by alt NHEJ is characterized by large deletions, resulting from extensive resection, and frequent chromosomal translocations (69,70). DNA ligase IIIα is the major enzyme acting in alt NHEJ but, surprisingly, this pathway does not appear to involve XRCC1 (71-73). There is also evidence for a minor DNA ligase I-dependent subpathway of alt NHEJ (71).
DNA ligase inhibitors
Attempts to identify DNA ligase inhibitors by screening of chemical libraries have been met with limited success (74-76). With the elucidation of the atomic resolution structure of human DNA ligase I complexed with nicked DNA in 2004 (4), it became possible to use a computer-aided drug design approach to identify DNA ligase inhibitors (Figure 4). A large database of small molecules was screened in silico for molecules that were predicted to bind in a pocket formed by residues His337, Arg449 and Gly453 on the DNA binding surface of the DBD of human DNA ligase I identified in the DNA ligase I-nicked DNA crystal structure (77,78). Of the approximately 200 compounds identified by the in silico screen, 10 inhibited human DNA ligase I but not T4 DNA ligase, an ATP-dependent bacteriophage DNA ligase that lacks a DBD. As expected based on the design of the screen, almost all of the compounds were competitive inhibitors with respect to nicked DNA (77,78). The exception was L82, which is an uncompetitive inhibitor of DNA ligase I. The inhibitors also exhibited activity against DNA ligases III and IV, presumably reflecting the conserved structure of the DBDs of the human DNA ligases. A DNA ligase I-specific inhibitor (L82) and inhibitors that inhibited DNA ligases I and III (L67) and all three DNA ligases (L189) were characterized further (77). These inhibitors retained their specificity in cell extract assays of DNA repair and also inhibited the growth of the human cells in culture. Although L67 and L189 are cytotoxic (77), the relative contributions of inhibition the function of DNA ligase IIIα in mitochondrial DNA replication and repair and inhibition of the function of both DNA ligase I and DNA ligase IIIα in nuclear DNA replication and repair to cell death have not been determined. Unexpectedly, subtoxic levels of L67 and L189 preferentially sensitized cancer cell lines to the cytotoxic effects of DNA agents (77), providing the first evidence that the DNA ligases inhibitors may have utility in the development of therapeutic strategies that target abnormalities in the DNA damage response of tumor cells.
In addition to their potential as cancer therapeutics, DNA ligase inhibitors can be used as probes to provide insights into the catalytic mechanisms and cellular functions of these enzymes but this will require the identification of more specific inhibitors. In a recent study, Srivatava et al. built a homology-based model of the DNA ligase IV DBD and used this to guide the selection of derivatives of L189 that may be more specific for DNA ligase IV (79). This screen identified a compound, SCR7, that inhibited DNA ligases III and IV but not DNA ligase I (79). Since atomic resolution structures are now available for DBDs of all the human DNA ligases (Figure 2B), more refined modeling approaches will likely lead to the identification of specific inhibitors of each of the human DNA ligases.
DNA ligases as biomarkers of abnormal DNA repair
The observation that DNA ligase inhibitors preferentially sensitize cancer cells to the cytotoxic effects of DNA damaging agents prompted an examination of the levels of human DNA ligases in cancer cell lines (77). As expected, the steady state levels of DNA ligase I were elevated in cancer cells lines compared with normal cells, presumably due to the increased proliferative activity of cancer cells (80). Strikingly, a pattern of elevated levels of DNA ligase IIIα and reduced levels of DNA ligase IV was observed in a significant fraction of cancer cell lines (77). Rassool and colleagues showed that this altered expression of DNA ligases IIIα and IV underlies the abnormal repair of DSBs in chronic myeloid leukemia (CML) cell lines expressing BCR-ABL1 (81,82). Specifically, the activity of DNA ligase IIIα-dependent alt NHEJ was significantly higher in these cell lines compared with normal myeloid cells. Interestingly, the steady levels of DNA ligases IIIα and PARP1, another alt NHEJ protein, and alt NHEJ activity itself were even higher in BCR-ABL1-postive CML cell lines that had acquired resistant to the ABL-specific inhibitor imatinib (82). A similar dysregulation of DSB repair occurs in cell lines expressing internal tandem duplication (ITD) mutations of the FMS-like tyrosine kinase-3 (FLT3) receptor that are present in a subset of acute myeloid leukemia patients with poor prognosis (83) and in breast cancer cell lines with acquired or intrinsic resistance to anti-estrogen therapies (84). Importantly, altered expression of DNA ligase IIIα and other proteins involved in the repair of DSBs by NHEJ have been detected in bone marrow samples from CML patients and tumor biopsies from breast cancer patients (82,84). Thus, the expression levels of DNA ligases IIIα and IV serve as biomarkers to identify cancers with abnormal repair of DSBs by alt NHEJ.
Activity of DNA ligase inhibitors in preclinical models of human cancer and patient samples
The increased activity of alt NHEJ in leukemia and breast cancer cell lines suggested that these cells may be preferentially sensitive to agents that inhibit alt NHEJ and induce DSBs. Notably, cell lines with increased alt NHEJ were hypersensitive to combination of a PARP inhibitor and the DNA ligase inhibitor L67 that inhibited alt NHEJ (82,84). Since knockdown of DNA ligase IIIα by RNAi had essentially the same effect as L67 in combination with the PARP inhibitor, it appears that L67 exerts its effect by inhibiting DNA ligase IIIα rather than DNA ligase I (82,84). Notably, studies with bone marrow samples from CML patients showed that sensitivity to the repair inhibitor combination correlated with the expression levels of both DNA ligase IIIα and PARP-1 (82). Thus, it appears that alt NHEJ is a valid therapeutic target in certain tyrosine kinase-activated leukemias and breast cancers and, more importantly, it appears to occur more frequently in forms of these diseases that are resistant to frontline therapies.
A recent study with SCR7, an inhibitor of DNA ligases IIIα and IV, suggested that the major DNA ligase IV-dependent NHEJ pathway may also be a therapeutic target in certain cancers (79). Specifically, SCR7 reduced cell proliferation in a DNA ligase IV-dependent manner and its effect on the growth of cancer cell lines appeared to correlate with the DNA ligase IV expression levels (79). In addition, SCR7 reduced the growth of tumors formed by several human cancer cell lines in mouse xenografts and, in tumors that did not respond to SCR7, the presence of SCR7 increased the tumor-inhibitory effects of agents that cause DSBs (79).
Conclusions
The requirement for DNA ligation in DNA replication and almost all DNA repair pathways makes DNA ligases an attractive target for the development of inhibitors that will potentiate the activity of genotoxic agents used to treat cancer. This effort is complicated by the presence of multiple species of DNA ligase in human cells with distinct but overlapping cellular functions and the essential roles of DNA ligases in the maintenance and replication of nuclear and mitochondrial genomes. Nonetheless, DNA ligase inhibitors identified by rational structure-based approaches have produced promising results in cell culture and mouse models of human cancer. In one of these examples, the DNA ligase inhibitor L67 was used in combination with a PARP inhibitor to selectively target a DNA repair abnormality in cancer cell lines and clinical samples from patients with leukemia that can be identified based upon altered expression of the human DNA ligases. Thus, the results of these preclinical studies support the further development of DNA ligase inhibitors as therapeutic agents for the selective targeting of DNA repair defects in cancer cells.
Acknowledgments
Funding: Studies on DNA ligases in the Tomkinson laboratory are supported by the Structural Cell Biology of DNA Repair Program Grant (P01 CA92584) and other research grants from the National Institutes of Health (GM47251, GM57479 and ES12512). A.E.T. is the Victor and Ruby Hansen Surface Endowed Chair in Cancer Research at the University of New Mexico Cancer Center.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David J. Chen and Benjamin P.C. Chen) for the series “DNA Damage and Repair” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Dr. Tomkinson is a co-inventor of a patent that covers the use of DNA ligase inhibitors as anticancer agents and a patent application that describes a method for detecting abnormalities in DNA double strand break repair in cancer cells. No potential conflicts of interest were declared by the other authors.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Friedberg EC, Walker GC, Siede W, et al. DNA Repair and Mutagenesis. 2nd ed. ASM Press, 2006.
- Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol 2008;8:363-9. [PubMed]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem 2008;77:313-38. [PubMed]
- Pascal JM, O’Brien PJ, Tomkinson AE, et al. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 2004;432:473-8. [PubMed]
- Tomkinson AE, Lasko DD, Daly G, et al. Mammalian DNA ligases. Catalytic domain and size of DNA ligase I. J Biol Chem 1990;265:12611-7. [PubMed]
- Cotner-Gohara E, Kim IK, Hammel M, et al. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry 2010;49:6165-76. [PubMed]
- De Ioannes P, Malu S, Cortes P, et al. Structural basis of DNA ligase IV-Artemis interaction in nonhomologous end-joining. Cell Rep 2012;2:1505-12. [PubMed]
- Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol 1999;19:3869-76. [PubMed]
- Mackey ZB, Ramos W, Levin DS, et al. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol Cell Biol 1997;17:989-98. [PubMed]
- Caldecott KW, McKeown CK, Tucker JD, et al. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol 1994;14:68-76. [PubMed]
- Dulic A, Bates PA, Zhang X, et al. BRCT domain interactions in the heterodimeric DNA repair protein XRCC1-DNA ligase III. Biochemistry 2001;40:5906-13. [PubMed]
- Nash RA, Caldecott KW, Barnes DE, et al. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 1997;36:5207-11. [PubMed]
- Cuneo MJ, Gabel SA, Krahn JM, et al. The structural basis for partitioning of the XRCC1/DNA ligase III-α BRCT-mediated dimer complexes. Nucleic Acids Res 2011;39:7816-27. [PubMed]
- Cotner-Gohara E, Kim IK, Tomkinson AE, et al. Two DNA-binding and nick recognition modules in human DNA ligase III. J Biol Chem 2008;283:10764-72. [PubMed]
- Mackey ZB, Niedergang C, Murcia JM, et al. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J Biol Chem 1999;274:21679-87. [PubMed]
- Taylor RM, Whitehouse J, Cappelli E, et al. Role of the DNA ligase III zinc finger in polynucleotide binding and ligation. Nucleic Acids Res 1998;26:4804-10. [PubMed]
- Grawunder U, Wilm M, Wu X, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 1997;388:492-5. [PubMed]
- Grawunder U, Zimmer D, Fugmann S, et al. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell 1998;2:477-84. [PubMed]
- Grawunder U, Zimmer D, Kulesza P, et al. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J Biol Chem 1998;273:24708-14. [PubMed]
- Doré AS, Furnham N, Davies OR, et al. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair (Amst) 2006;5:362-8. [PubMed]
- Sibanda BL, Critchlow SE, Begun J, et al. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol 2001;8:1015-9. [PubMed]
- Wu PY, Frit P, Meesala S, et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 2009;29:3163-72. [PubMed]
- Levin DS, Bai W, Yao N, et al. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci U S A 1997;94:12863-8. [PubMed]
- Montecucco A, Rossi R, Levin DS, et al. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J 1998;17:3786-95. [PubMed]
- Montecucco A, Savini E, Weighardt F, et al. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J 1995;14:5379-86. [PubMed]
- Howes TR, Tomkinson AE. DNA ligase I, the replicative DNA ligase. Subcell Biochem 2012;62:327-41. [PubMed]
- Vijayakumar S, Dziegielewska B, Levin DS, et al. Phosphorylation of human DNA ligase I regulates its interaction with replication factor C and its participation in DNA replication and DNA repair. Mol Cell Biol 2009;29:2042-52. [PubMed]
- Levin DS, McKenna AE, Motycka TA, et al. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol 2000;10:919-22. [PubMed]
- Barnes DE, Tomkinson AE, Lehmann AR, et al. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell 1992;69:495-503. [PubMed]
- Henderson LM, Arlett CF, Harcourt SA, et al. Cells from an immunodeficient patient (46BR) with a defect in DNA ligation are hypomutable but hypersensitive to the induction of sister chromatid exchanges. Proc Natl Acad Sci U S A 1985;82:2044-8. [PubMed]
- Bentley D, Selfridge J, Millar JK, et al. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet 1996;13:489-91. [PubMed]
- Bentley DJ, Harrison C, Ketchen AM, et al. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J Cell Sci 2002;115:1551-61. [PubMed]
- Le Chalony C, Hoffschir F, Gauthier LR, et al. Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell Mol Life Sci 2012;69:2933-49. [PubMed]
- Arakawa H, Bednar T, Wang M, et al. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res 2012;40:2599-610. [PubMed]
- Caldecott KW, Tucker JD, Stanker LH, et al. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res 1995;23:4836-43. [PubMed]
- Okano S, Lan L, Caldecott KW, et al. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 2003;23:3974-81. [PubMed]
- Tebbs RS, Flannery ML, Meneses JJ, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol 1999;208:513-29. [PubMed]
- Puebla-Osorio N, Lacey DB, Alt FW, et al. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol 2006;26:3935-41. [PubMed]
- Gao Y, Katyal S, Lee Y, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature 2011;471:240-4. [PubMed]
- Simsek D, Furda A, Gao Y, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 2011;471:245-8. [PubMed]
- Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res 2000;28:3880-6. [PubMed]
- Lakshmipathy U, Campbell C. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res 2001;29:668-76. [PubMed]
- Frosina G, Fortini P, Rossi O, et al. Two pathways for base excision repair in mammalian cells. J Biol Chem 1996;271:9573-8. [PubMed]
- Kubota Y, Nash RA, Klungland A, et al. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J 1996;15:6662-70. [PubMed]
- Matsumoto Y, Kim K, Hurwitz J, et al. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J Biol Chem 1999;274:33703-8. [PubMed]
- Teo IA, Arlett CF, Harcourt SA, et al. Multiple hypersensitivity to mutagens in a cell strain (46BR) derived from a patient with immuno-deficiencies. Mutat Res 1983;107:371-86. [PubMed]
- Teo IA, Broughton BC, Day RS, et al. A biochemical defect in the repair of alkylated DNA in cells from an immunodeficient patient (46BR). Carcinogenesis 1983;4:559-64. [PubMed]
- Harrison C, Ketchen AM, Redhead NJ, et al. Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer Res 2002;62:4065-74. [PubMed]
- Aboussekhra A, Biggerstaff M, Shivji MK, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 1995;80:859-68. [PubMed]
- Araújo SJ, Tirode F, Coin F, et al. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev 2000;14:349-59. [PubMed]
- Moser J, Kool H, Giakzidis I, et al. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell 2007;27:311-23. [PubMed]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 2008;9:619-31. [PubMed]
- Amé JC, Rolli V, Schreiber V, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 1999;274:17860-8. [PubMed]
- Ménissier de Murcia J, Ricoul M, Tartier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 2003;22:2255-63. [PubMed]
- Okano S, Lan L, Tomkinson AE, et al. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res 2005;33:422-9. [PubMed]
- Katyal S, McKinnon PJ. Disconnecting XRCC1 and DNA ligase III. Cell Cycle 2011;10:2269-75. [PubMed]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010;79:181-211. [PubMed]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008;77:229-57. [PubMed]
- DeFazio LG, Stansel RM, Griffith JD, et al. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 2002;21:3192-200. [PubMed]
- Girard PM, Kysela B, Härer CJ, et al. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum Mol Genet 2004;13:2369-76. [PubMed]
- O’Driscoll M, Cerosaletti KM, Girard PM, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell 2001;8:1175-85. [PubMed]
- Riballo E, Critchlow SE, Teo SH, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol 1999;9:699-702. [PubMed]
- Barnes DE, Stamp G, Rosewell I, et al. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol 1998;8:1395-8. [PubMed]
- Frank KM, Sekiguchi JM, Seidl KJ, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 1998;396:173-7. [PubMed]
- Corneo B, Wendland RL, Deriano L, et al. Rag mutations reveal robust alternative end joining. Nature 2007;449:483-6. [PubMed]
- Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 2007;449:478-82. [PubMed]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 2010;17:410-6. [PubMed]
- Fattah F, Lee EH, Weisensel N, et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 2010;6:e1000855 [PubMed]
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell 2007;131:223-5. [PubMed]
- Lieber MR. NHEJ and its backup pathways in chromosomal translocations. Nat Struct Mol Biol 2010;17:393-5. [PubMed]
- Simsek D, Brunet E, Wong SY, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 2011;7:e1002080 [PubMed]
- Wang H, Rosidi B, Perrault R, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 2005;65:4020-30. [PubMed]
- Boboila C, Oksenych V, Gostissa M, et al. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc Natl Acad Sci U S A 2012;109:2473-8. [PubMed]
- Sun D, Urrabaz R. Development of non-electrophoretic assay method for DNA ligases and its application to screening of chemical inhibitors of DNA ligase I. J Biochem Biophys Methods 2004;59:49-59. [PubMed]
- Tan GT, Lee S, Lee IS, et al. Natural-product inhibitors of human DNA ligase I. Biochem J 1996;314:993-1000. [PubMed]
- Tseng HM, Shum D, Bhinder B, et al. A high-throughput scintillation proximity-based assay for human DNA ligase IV. Assay Drug Dev Technol 2012;10:235-49. [PubMed]
- Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 2008;68:3169-77. [PubMed]
- Zhong S, Chen X, Zhu X, et al. Identification and validation of human DNA ligase inhibitors using computer-aided drug design. J Med Chem 2008;51:4553-62. [PubMed]
- Srivastava M, Nambiar M, Sharma S, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 2012;151:1474-87. [PubMed]
- Sun D, Urrabaz R, Nguyen M, et al. Elevated expression of DNA ligase I in human cancers. Clin Cancer Res 2001;7:4143-8. [PubMed]
- Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood 2008;112:1413-23. [PubMed]
- Tobin LA, Robert C, Rapoport AP, et al. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene 2013;32:1784-93. [PubMed]
- Fan J, Li L, Small D, et al. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood 2010;116:5298-305. [PubMed]
- Tobin LA, Robert C, Nagaria P, et al. Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res 2012;10:96-107. [PubMed]


