
Relevance of the Hedgehog pathway in T-cell acute lymphoblastic leukemia
T-cell lymphoblastic leukemias (T-ALLs) arise from the malignant transformation of T cell progenitors. The most common genetic alterations include the activation of the NOTCH pathway, deletions within the CDKN2A locus and chromosomal rearrangements affecting T cell-specific enhancer elements. In addition, alterations in other signal transductions pathways and chromatin remodelers translate into the genetic heterogeneity of the disease. T-ALL accounts for 10%–15% of pediatric and 25% of adult cases. Clinical symptoms include hyperleukocytosis with extramedullary involvement of lymph nodes and other organs, including frequent central nervous system infiltration and occurrence of mediastinal masses arising from the thymus. The outcome of T-ALL patients has improved due to intensified chemotherapy protocols and cure rates of 75% in children and 50% in adults can be achieved. But the prognosis of primary resistant or relapsed T-ALL patients remains poor underlining the need for novel targeted therapy strategies (1).
Many studies focus on the Hedgehog (HH) pathway as suitable target for directed cancer therapies. The Hedgehog pathway is involved in stem cell proliferation, differentiation, and survival. In mammals, the classical canonical Hedgehog pathway can be activated through three different ligands: Sonic Hedgehog (SHH), Desert Hedgehog (DHH), and Indian Hedgehog (IHH), which bind to the transmembrane receptor Patched-1 (PTCH1). Without the presence of ligands, PTCH1 represses activation of the transmembrane protein Smoothened (SMO). Upon HH ligand binding, PTCH1 is internalized, resulting in relief and thus activation of SMO that transduces the signal to the family of Glioma (GLI) zinc finger transcription factors GLI1, GLI2, and GLI3. GLI1 and GLI2 predominantly represent transcriptional activators while GLI3 acts as transcriptional repressor. Furthermore, Suppressor of Fused (SUFU) acts as negative pathway regulator by inhibiting the activation of GLI transcription factors in the absence of HH ligands (2). In addition to this classical canonical signaling axis, different mechanisms of non-canonical pathway activation have been described leading to activation of the GLI transcription factors independent of Hedgehog ligands or PTCH/SMO. Non-canonical HH pathway activation includes Ras-ERK, TGF-β, or PI3K-AKT (3-5).
Aberrant Hedgehog signaling has been described in a broad variety of cancers. Some cancers such as basal cell carcinoma or medulloblastoma show high frequencies of somatic mutations in PTCH1, SMO or SUFU that account for the dysregulation of the pathway. In other cancers, mutations in Hedgehog family members occur only at low frequencies and different mechanisms including enhanced autocrine or paracrine as well as non-canonical pathway activation might account for the dysregulated HH signaling in those cancers (5). The expression of Hedgehog family members could be correlated to the patients’ outcome in a number of cancers such as colon, bladder and breast cancer or acute myeloid leukemia (AML) (3,6,7). The therapeutic potential of the HH pathway inhibition was demonstrated in many preclinical studies that led to the evaluation in the clinical setting. So far, the small molecule SMO inhibitors GDC-0449 (vismodegib) and LDE225 (sonidegib) have been approved for the treatment of advanced basal cell carcinoma. Furthermore, a multiplicity of clinical trials is ongoing investigating GDC-0449, LDE225 and a number of other SMO inhibitors such as glasdegib, BMS-833923, taladegib or saridegib in many solid and hematological cancers (5).
The importance of HH signaling in healthy adult as well as malignant hematopoiesis is discussed controversially. While described as dispensable in some studies (8,9), recent data suggest the involvement of the HH signaling cascade in different types of leukemia implying that the HH pathway might represent a valuable therapeutic target (7,10-12). Based on promising preclinical data, first clinical studies investigating the therapeutic potential of HH inhibitors in hematological malignancies are ongoing. Encouraging data from a phase II randomized trial of AML and myelodysplastic syndrome (MDS) patients treated with the SMO inhibitor glasdegib were recently presented at the ASH Annual Meeting (13). Several clinical trials using the SMO inhibitor LDE225 are ongoing including a phase I study combining LDE225 with azacitidine or decitabine in myeloid malignancies (NCT02129101). A phase II study with LDE225 in acute leukemia and a phase I study with LDE225 in combination with nilotinib in CML have already been completed but the results are still awaited (NCT01826214 and NCT01456676, respectively) while, disappointingly, a phase II study with GDC-0449 in patients with AML and MDS had to be prematurely terminated due to lower-than-expected efficacy (NCT01880437).
The Hedgehog pathway is essential for normal T-cell development as was shown using mice with defects in HH family members. Mice lacking PTCH are characterized by a severely affected T-cell development resulting in thymic atrophy and SHH knockout mice have significantly reduced thymocyte levels during development (14,15).
In a recently published preclinical study, Dagklis and colleagues provide evidence that the HH pathway is activated a subgroup of T-cell acute lymphoblastic leukemia patients and that those patients might benefit from treatment with HH inhibitors (16).
In detail, the authors investigated the mRNA expression of HH family members in two independent T-ALL patient cohorts [RNA sequencing data and published Affymetrix arrays, n=51 and n=88, respectively; (17)] and found ectopic expression of SHH, IHH and GLI1 in 22% of T-ALL cases. The expression of GLI1 was strongly correlated to the expression of the HH ligands indicating to an autocrine stimulation loop in those T-ALL samples. Furthermore, the expression of HH pathway transcripts was also significantly higher in T-ALL samples in comparison to normal T-cell subsets. Interestingly, the HH pathway activation in T-ALL patients could not be linked to any specific T-ALL subtype but seems to represent an independent subgroup. The authors further analyzed their data using the computational method iRegulon in order identify transcriptional regulatory networks. Using this approach, they revealed a strong correlation between the expression of GLI1 and a number of transcription factors involved in leukemogenesis.
Furthermore, Dagklis and colleagues investigated the impact of Hedgehog signaling in a T-ALL mouse model. They chose their previously published JAK3(M511I) T-ALL mouse model as they observed a strong correlation between the expression of GLI1 and JAK3 in human T-ALL samples and suggested that both pathways might synergize with regard to leukemia development. The combined co-expression of SHH or IHH with JAK3(M511I) had no significant influence on the disease latency. Nevertheless, the overexpression of HH components had a clear impact on the leukemia development as those clones expressing both the HH ligand and the JAK3 mutant had a clonal advantage compared to clones expressing only one of the components. Indeed, leukemic cells expressing both the ligand and JAK3(M511I) became the dominant clone in all hematopoietic organs and had higher lymph node infiltration rates. These data seem to be contradictory to data published by Gao et al. as they found no impact of the HH pathway in a murine T-ALL model (8). Both research groups might be right as there are key differences between both mouse models. Dagklis and colleagues combined overexpression of HH ligand to a JAK3 mutation model with a long latency. Whereas on the other hand, Gao and colleagues used the NOTCH-ICN model which was combined with a SMO deficiency in order to investigate the role of HH signaling with regard to T-ALL. The NOTCH-ICN mouse model is quite aggressive with a latency of only about 20 days until the mice develop a leukemic phenotype which might explain why no difference could be observed in this study. These data reveal the complexity and context-dependency of the process of cancer biology and should prevent us from drawing hasty conclusions.
Dagklis and colleagues further observed that also non-leukemic cells were affected by the activated HH cascade in their mouse model. They could detect higher GLI1 levels in normal T cells and thymic epithelial cells (TECs) which are part of the stromal thymic cells. Interestingly, the TECs showed increased expression of Delta-like 4 (Dll4), Interleukin-7 (IL7) and Vascular Endothelial Growth Factor. In previous studies, it was shown that murine and human primary T-ALL samples require stromal-derived signals such as NOTCH ligands or IL7 for their survival. Therefore the authors proposed that T-ALL cells and TECs might interact in a paracrine manner: the leukemic cells secrete HH ligands that stimulate TECs in the thymus which in turn secrete prosurvival factors for the leukemic cells.
Hedgehog-mediated paracrine interactions between tumor and stroma cells represent an accepted concept in cancer biology. Secretion of HH ligands by tumor cells leading to the HH cascade activation in the tumor stroma which in turn provide protumoral growth factors has been described in many cancer entities (3,4,18). On the other hand, paracrine activation of tumor cells through stromal-derived HH ligands was also observed in several hematological cancers such as AML and CLL (7,10,11,19). For instance, Hedge and colleagues showed that stroma-induced HH signaling could increase the survival of B-cell chronic lymphocytic leukemia (B-CLL) cells, whereas treatment of the coculture with the HH inhibitor cyclopamine abrogated the stroma-induced prosurvival effects. Furthermore, gene expression profiling of primary B-CLL cells revealed that HH signaling molecules, including GLI1 and GLI2, were increased and correlated with the patients' disease progression (11). Besides, we could recently show that stromal cells contribute to the activated HH signaling in AML by provision of DHH (7).
Dagklis et al. evaluated the potential of a HH pathway blockade in vitro using 9 different T-ALL cell lines. Cells were treated with the SMO inhibitors cyclopamine or GDC-0449 or the GLI inhibitor GANT61. Cyclopamine and GDC-0449 caused a dose-dependent inhibition of proliferation in three out of nine analyzed cell lines while GANT61 was more effective and reduced the cell growth in six out of nine T-ALL cell lines. As already mentioned above, the HH pathway can be activated both in a canonical as well as non-canonical manner (3-5). These data show that the activation of the GLI transcription factors seems to be SMO-independent in a part of T-ALL cells as the inhibition of SMO had no effect in contrast to a direct inhibition of the HH downstream mediator GLI. Several GLI activating signaling cascades have been described including Ras-ERK, TGF-β, or PI3K-AKT (3-5,20). In addition to an autocrine DHH-SMO axis, Decker and colleagues identified a SMO-independent non-canoncial activation of the HH cascade through ERK signaling in CLL cells (3). In our previous work, we observed an association between the occurrence of FLT3 mutations and high GLI2 expressions levels in AML patients indicating at a non- canonical activation mechanisms of GLI trough FLT3 and its downstream target PI3K (7). Furthermore, the combination of a FLT3 inhibitor and GANT61 as direct GLI inhibitor resulted in significantly increased anti-leukemic effects in FLT3-mutated AML cells compared to single agent treatments [(20) and unpublished data]. These data are in line with a study by Lim and colleagues as they observed a shortened AML latency upon combination of FLT3-ITD and with a constitutively active SMO in a leukemic mouse model (21). The PI3K cascade might also represent a potential candidate for the non-canonical HH pathway activation in T-ALL as 5–10% of T-ALL patients harbor deletion mutations of the tumor suppressor PTEN which is a critical negative regulator of PI3K signaling. Whether PI3K, one of the other common or further signaling cascades contribute to the non-canonical HH activation in T-ALL should be investigated in future studies (1). Possible mechanisms of the Hedgehog pathway activation are summarized in Figure 1.
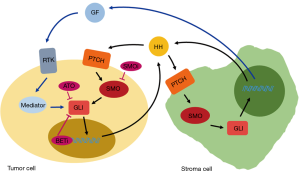
What have we learned from recently published studies about the Hedgehog pathway in cancer? Its central role due to activating mutations in cancers such as basal cell carcinoma or medulloblastoma is known for years now and HH inhibitors already found their way into the clinic. But recent data show that dysregulated HH signaling is not restricted to a limited number of cancer entities. Indeed, it seems to be of crucial importance in nearly every cancer including many types of hematological malignancies which makes it a very promising therapeutic target. But: the HH signaling cascade is far more complex than the classical canonical activation axis via HH ligands, PTCH1, SMO and the GLI transcription factors. On the one hand, the signaling cascade is often activated through paracrine interactions of tumor and stromal cells that involve a multitude of other signaling networks. On the other hand, more and more studies reveal the prominent role of the non-canonical HH signaling cascade in cancer. There seems to be a wide variety of possible SMO-independent activation mechanisms indicating that the direct inhibition of GLI might represent the preferentially therapeutic target in comparison to SMO. Although quite effective in preclinical models, GLI inhibitors such as GANT58 or GANT61 are unlikely to be clinically evaluated due to their high in vitro und in vivo IC50 values. But at least two other options to inhibit GLI in the clinical setting are available: arsenic trioxide (ATO) and Bromodomain and extraterminal domain (BET) protein inhibitors.
ATO is FDA-approved for the treatment of acute promyelocytic leukemia (APL) due to its binding to the malignant fusion protein PML-RARA and its following degradation (22). But ATO has also been described as direct GLI inhibitor that improved the survival in a murine medulloblastoma model with activated HH pathway signaling (23). Although not primarily tested as GLI inhibitor, ATO is currently evaluated as therapeutic target in a number of hematological malignancies including in addition to APL also AML, CML or MDS (e.g., NCT02339740, NCT02200978, NCT02188706, NCT02190695, NCT02188706).
BET proteins are epigenetic modulators regulating gene transcription through recruitment of regulatory complexes to acetylated chromatin regions (24). BRD4, the most prominent family member, was shown to regulate the transcription of important proto-oncogenes. Interestingly, BRD4 was recently identified as regulator of GLI1 and GLI2 (25). Preclinical data revealed the potential of BRD4 inhibitors in hematological malignancies (26,27), that could be strengthened by first clinical results in AML (28). As part of the observed effects is likely to origin from the inhibition of GLI, BRD4 inhibitors should be considered as treatment approach in hematological malignancies including the subgroup of T-ALL patients with an activated HH signaling cascade.
Taken together, patients with hematological malignancies including the subgroup of T-ALL patients with an activated HH signaling cascade might benefit from HH-targeting therapies. Due to the growing evidence of SMO-independent non-canonical HH pathway activation in cancer, GLI inhibitors such as ATO or BRD4 inhibitors should be the substances of choice.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peipei Xu (Department of Hematology, the Affiliated Drum Tower Hospital of Nanjing University Medical School, Nanjing, China).
Conflicts of Interest: JW and WF: Inventorship on patents held by Amgen Research (Munich) GmbH/Amgen Inc., WF: Travel grant, advisory board and research funding by Amgen Inc., travel grant and advisory board by TEVA GmbH, advisory board: Ariad/Incyte Inc., travel grant by Gilead Inc., research funding by Pfizer Inc.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 2012;122:3398-406. [Crossref] [PubMed]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature 2001;411:349-54. [Crossref] [PubMed]
- Decker S, Zirlik K, Djebatchie L, et al. Trisomy 12 and elevated GLI1 and PTCH1 transcript levels are biomarkers for Hedgehog-inhibitor responsiveness in CLL. Blood 2012;119:997-1007. [Crossref] [PubMed]
- Kebenko M, Drenckhan A, Gros SJ, et al. ErbB2 signaling activates the Hedgehog pathway via PI3K-Akt in human esophageal adenocarcinoma: identification of novel targets for concerted therapy concepts. Cell Signal 2015;27:373-81. [Crossref] [PubMed]
- Rimkus TK, Carpenter RL, Qasem S, et al. Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers (Basel) 2016;8. [PubMed]
- Gonnissen A, Isebaert S, Haustermans K. Targeting the Hedgehog signaling pathway in cancer: beyond Smoothened. Oncotarget 2015;6:13899-913. [Crossref] [PubMed]
- Wellbrock J, Latuske E, Köhler J, et al. Expression of Hedgehog Pathway Mediator GLI Represents a Negative Prognostic Marker in Human Acute Myeloid Leukemia and Its Inhibition Exerts Antileukemic Effects. Clin Cancer Res 2015;21:2388-98. [Crossref] [PubMed]
- Gao J, Graves S, Koch U, et al. Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell 2009;4:548-58. [Crossref] [PubMed]
- Hofmann I, Stover EH, Cullen DE, et al. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell 2009;4:559-67. [Crossref] [PubMed]
- Dierks C, Beigi R, Guo GR, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell 2008;14:238-49. [Crossref] [PubMed]
- Hegde GV, Peterson KJ, Emanuel K, et al. Hedgehog-induced survival of B-cell chronic lymphocytic leukemia cells in a stromal cell microenvironment: a potential new therapeutic target. Mol Cancer Res 2008;6:1928-36. [Crossref] [PubMed]
- Hou X, Chen X, Zhang P, et al. Inhibition of hedgehog signaling by GANT58 induces apoptosis and shows synergistic antitumor activity with AKT inhibitor in acute T cell leukemia cells. Biochimie 2014;101:50-9. [Crossref] [PubMed]
- Cortes JE, Heidel FH, Heuser M, et al. A phase 2 randomized study of low dose Ara-C with or without glasdegib (PF- 04449913) in untreated patients with acute myeloid leukemia or high-risk myelodysplastic syndrome. ASH Annual Meeting Abstracts, 2016.
- Shah DK, Hager-Theodorides AL, Outram SV, et al. Reduced thymocyte development in sonic hedgehog knockout embryos. J Immunol 2004;172:2296-306. [Crossref] [PubMed]
- Uhmann A, Dittmann K, Nitzki F, et al. The Hedgehog receptor Patched controls lymphoid lineage commitment. Blood 2007;110:1814-23. [Crossref] [PubMed]
- Dagklis A, Demeyer S, De Bie J, et al. Hedgehog pathway activation in T-cell acute lymphoblastic leukemia predicts response to SMO and GLI1 inhibitors. Blood 2016;128:2642-54. [Crossref] [PubMed]
- Atak ZK, Gianfelici V, Hulselmans G, et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet 2013;9:e1003997 [Crossref] [PubMed]
- Heller E, Hurchla MA, Xiang J, et al. Hedgehog signaling inhibition blocks growth of resistant tumors through effects on tumor microenvironment. Cancer Res 2012;72:897-907. [Crossref] [PubMed]
- Zou J, Hong Y, Tong Y, et al. Sonic hedgehog produced by bone marrow-derived mesenchymal stromal cells supports cell survival in myelodysplastic syndrome. Stem Cells Int 2015;2015:957502.
- Latuske E, Stamm H, Klokow M, et al. Combined inhibition of Hedgehog and FLT3 signalling leads to effective anti- leukemic effects in acute myeloid leukemia. EHA Annual Meeting Abstract, 2015.
- Lim Y, Gondek L, Li L, et al. Integration of Hedgehog and mutant FLT3 signaling in myeloid leukemia. Sci Transl Med 2015;7:291ra96 [Crossref] [PubMed]
- Zeidan AM, Gore SD. New strategies in acute promyelocytic leukemia: moving to an entirely oral, chemotherapy-free upfront management approach. Clin Cancer Res 2014;20:4985-93. [Crossref] [PubMed]
- Beauchamp EM, Ringer L, Bulut G, et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest 2011;121:148-60. [Crossref] [PubMed]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014;54:728-36. [Crossref] [PubMed]
- Tang Y, Gholamin S, Schubert S, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 2014;20:732-40. [Crossref] [PubMed]
- Da Costa D, Agathanggelou A, Perry T, et al. BET inhibition as a single or combined therapeutic approach in primary paediatric B-precursor acute lymphoblastic leukaemia. Blood Cancer J 2013;3:e126 [Crossref] [PubMed]
- Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524-8. [Crossref] [PubMed]
- Berthon C, Raffoux E, Thomas X, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol 2016;3:e186-95. [Crossref] [PubMed]


