Molecular resistance mechanisms of ALK inhibitors and implications for therapeutic management of ALK-rearranged lung cancer patients
Dysregulated anaplastic lymphoma kinase (ALK) protein expression has been previously reported in non-Hodgkin’s lymphoma (NHL) (1). However, it was not until 2007 when Soda et al. and others revealed that ALK is constitutively activated in some patients with non-small cell lung cancer (NSCLC), due to ALK gene rearrangement (2,3). In NSCLC, ALK rearrangement results in expression of ALK fusion proteins with aberrant ALK signalling and oncogenic transformation (2) and occurs in about 3–5% of the total NSCLCs (4). Currently, the treatment strategy of so-called ALK rearranged NSCLC relies on selection of an ALK tyrosine kinase inhibitor (TKI). The first in class ALK inhibitor crizotinib was developed and approved through accelerated drug approval by the US Food and Drug Administration (FDA) in 2011 on the basis of high response rates in early phase evaluation (5,6) and was granted regular approval by US FDA in 2013 based on demonstration of superior progression-free survival (PFS) and overall response rate (RR) for crizotinib-treated patients compared to chemotherapy (7). However, despite the significant improvements observed in ALK-rearranged NSCLC patients with crizotinib compared to conventional cytotoxic chemotherapy, resistance to crizotinib occurs with patients often relapsing within 1–2 years (8-10). Several second-generation ALK inhibitors have been developed and tested in patients who have progressed on treatment with crizotinib (11,12). Among these, ceritinib and alectinib were recently approved by the FDA for treatment of crizotinib-resistant ALK-rearranged NSCLC patients (13-15) and have also demonstrated clinical efficacy in crizotinib naïve patients (16,17) but resistance also occurs to these drugs (18,19). The question then is how best to select and schedule ALK inhibitors to optimise treatment for ALK rearranged NSCLC? To address this requires knowledge of ALK TKI resistance mechanisms and how this knowledge can be applied in the clinic. A recent study conducted by Gainor and colleagues from the group of Dr. Alice T Shaw provides new insights into ALK TKI resistance mechanisms that has implications for ALK treatment selection in the clinic (20).
Generally, resistance mechanisms to ALK TKIs can be classified into two main categories, including on-target genetic modifications such as ALK resistance mutations or ALK gene amplification and off-target changes including dysregulation of bypass signalling molecules to compromise ALK inhibition by ALK TKIs. The off-target resistance mechanisms remain poorly understood but currently include mutation of several key signalling molecules such as EGFR and KRAS and activation of pro-survival signalling pathways and hypoxia-induced epithelial-mesenchymal transition (EMT) (21). Consequently, combinatorial strategies to target ALK alongside an off-target resistance mechanism are being tested. For example, crizotinib and imatinib (KIT-TKI) or OSI-906 (IGF-1R-TKI) respectively for bypass signal-induced resistance to crizotinib mediated by X-376 of the KIT and IGF-1R pathways (10,22). To date most progress has been made in characterising on-target mechanisms that account for about 33% of total crizotinib resistance in ALK rearranged NSCLC patients (10). Several secondary mutations within the ALK gene in response to targeted treatment with either crizotinib or the second-generation of ALK inhibitors ceritinib and alectinib have been identified. At least 11, 3 and 6 different mutations have been reported to emerge during treatment with crizotinib, alectinib and ceritinib respectively (21) with some mutations such as ALK G1202R conferring resistance to crizotinib, ceritinib and alectinib (11,19).
Dr. Gainor and colleagues analysed 103 repeat biopsies from 83 patients with ALK-rearranged NSCLC who had progressed on treatment with one or more ALK inhibitors (20). Their results demonstrate a distinct spectrum of ALK resistance mutations for different ALK inhibitors and that mutations are more frequent following exposure to second generation ALK inhibitors. In this study crizotinib resistance mutations were confirmed in 11 (20%, N=51) specimens from biopsy sites that included pleural fluid (31%), liver (22%), and nodal tissue (18%) from 10 patients. L1196M and G1269A mutations represented the most common mutations with a frequency of 7% and 4% of the total mutations detected, respectively. Other mutations (frequency) detected were C1156Y (2%), G1202R (2%), I1171T (2%), S1206Y (2%), and E1210K (2%). Within 36 crizotinib-resistant specimens pre-confirmed by ALK FISH testing as ALK rearranged tumours, 31% of the examined specimens were found to have on-target genetic alterations contributing to the crizotinib resistance and 3 (8.3%) specimens demonstrated ALK gene amplification. Interestingly, no resistant mutations were concomitant with the ALK gene amplification. Changes in ALK resistance mutational profile following treatment with the second-generation ALK inhibitors ceritinib (N=23), alectinib (N=17), or brigatinib (N=6) were also examined. The results obtained indicate that within the available specimens of 9 cases of pre-ceritinib/post-crizotinib only 2 (22%) exhibited on-target resistance mechanisms, including ALK resistance mutation S1206Y and ALK fusion gene amplification. Of 24 separate post-ceritinib biopsies (obtained from 23 ceritinib treated patients), 54% harbored ALK mutations, with 17% of the total ALK mutations exhibiting two different mutations concomitantly. The G1202R (21%) and F1174C/L (16.7%) mutations were most common. Of note, a novel ALK G1202del mutation was also identified in 8% of specimens. The authors conducted preclinical studies to determine the functional consequences of the various mutations identified in the clinical cases. Ectopic expression of the EML4–ALK G1202del mutant in Ba/F3 cells suggested that the G1202del ALK mutant confers moderate resistance to ceritinib, alectinib, and brigatinib with crizotinib potency being less affected. For 17 patients treated with alectinib (who had previously received crizotinib), 17 alectinib-resistant biopsies were analysed and ALK resistance mutations were detected in 9 (53%) specimens. Interestingly, the ALK G1202R mutation was present in 29% of cases. Detection of the ALK V1180L mutation (6%) in response to alectinib was confirmed for the first time in an alectinib-resistant patient. Finally, for the 7 patients treated with brigatinib, ALK resistance mutations were observed in 5 of 7 (71%), where the ALK G1202R mutant was detected in three specimens (60%).
Overall, these data indicate that patients treated with second generation ALK inhibitors compared to crizotinib as a first generation inhibitor have a higher frequency of ALK mutations with the higher resistance conferred by the ALK G1202R mutation also representing the most common detected mutation. The findings demonstrate that part of the mechanism by which dysregulated ALK tumours adapt to resist treatment with ALK TKIs relies on the potency of ALK inhibition. On exposure to a less potent ALK inhibitor (crizotinib) the ALK mutations that emerge exhibit moderate resistance capacity. However, the more potent second-generation ALK inhibitors are associated with a higher frequency of mutations with higher resistance capacity such as G1202R. However, it is unclear whether the mutation profile that emerges on treatment with a second generation ALK TKI is contributed to by prior exposure to crizotinib. Thus, the question that remains to be answered is whether patients with ALK rearranged NSCLC should be treated with second or third-generation ALK inhibitors as first line treatment or whether sequencing treatment as 1st, 2nd and 3rd generation inhibitors would be more beneficial. A recent pre-planned interim analysis from the J-ALEX clinical trial presented at the 2016 American Society of Clinical Oncology (ASCO) Annual meeting demonstrated prolonged PFS for treatment with alectinib compared to crizotinib in untreated ALK rearranged NSCLC patients (median PFS not reached versus 10.2 months; HR: 0.34; P<0.0001) (16), and the final data evaluating overall survival (OS) might provide more information about which is the better sequencing treatment strategy when the patients in the “crizotinib treatment arm” are crossed over to alectinib treatment when disease progresses. Furthermore, lorlatinib a third-generation ALK inhibitor has been found to be more effective in a number of patient-derived ceritinib-resistant cell lines harbouring ALK mutations (20). In another study by Shaw et al., a patient with metastatic ALK rearranged lung cancer developed crizotinib resistance due to a C1156Y ALK mutation. The patient did not respond to a second-generation ALK inhibitor, but responded to the third generation ALK inhibitor, lorlatinib before tumour relapse (23). Sequencing analysis revealed a further lorlatinib resistant ALK L1198F mutation in addition to C1156Y. The L1198F mutation was found to enhance the ALK crizotinib binding affinity within ALK and re-sensitise the tumour to crizotinib (23). Therefore, the usage of second or the third-generation inhibitors as first line treatment in ALK rearranged NSCLC patients could be an option. A further consideration for ALK inhibitor selection is the type of ALK rearrangement and fusion gene. Recent data published by Yoshida et al., that compared the differential crizotinib response duration among ALK fusion variants established using RT-PCR demonstrated ALK fusion variant 1 to be the most frequently detected variant (54%) (24). The objective response rate (ORR) was 74% and 63% in the variant 1 and non-variant 1 groups, respectively and 69% overall. The median PFS was significantly longer in patients with variant 1, 11.0 (95% CI, 6.5–43.0) months than in those with non-variant 1, 4.2 (95% CI, 1.6–10.2) months, respectively (P<0.05) (24). Further investigations evaluating survival endpoints for the different specific types of ALK fusions, and for the most common mutations, such as L1196M, G1269A, C1156Y, G1202R, I1171T, S1206Y, and E1210K would be of value to develop optimal clinical algorithms for ALK inhibitor selection.
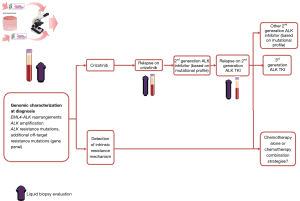
From the aforementioned, it is clear that detection of ALK TKI resistance mutations are a critical point in determining treatment strategy, although not yet fully integrated into routine practice. Repeat biopsy and sequencing analysis to assess for the presence and type of resistance mutation following treatment with an ALK TKI seems set to become the norm. However, tissue biopsy specimens are always limited to certain locations and hardly reflect the comprehensive molecular signatures of metastatic lung cancer, and intratumour heterogeneity. In contrast liquid biopsies can be obtained from almost all body fluids, thus representing a new source of cancer-derived materials to better reflect the nature of tumour at both primary and metastatic sites. Detection of circulating tumour cells (CTCs), circulating tumour DNA (ctDNA), circulating tumour RNA (ctRNA), exosomes, and tumour-educated platelets (TEPs) in body fluids samples have potential to provide much more information regarding ALK resistance mechanisms compared with tissue biopsies. However, due to the lower frequency of these markers in body fluids, achieving higher detection sensitivity and specificity remains technically challenging (25). Table 1 summaries the current liquid biopsy strategies for ALK resistance evaluation with the observed limitations. Evaluation of intrinsic or acquired TKI resistance alterations in patients with ALK rearranged NSCLC using liquid biopsy can direct treatment selection based on a time specific mutational profile and anticipate treatment resistance to second generation ALK inhibitors. Additionally, liquid biopsy might overcome the tumour heterogeneity limitations of tissue biopsies allowing for the detection of acquired compound mutations associated to specific TKI resistance (Figure 1).
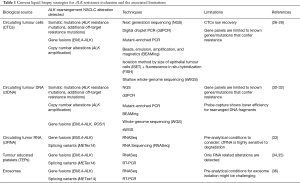
Full table
In summary, despite clinical studies that demonstrate better RR and benefits of ALK TKIs in patients with ALK rearranged NSCLC, resistance to ALK TKIs remains challenging. Better knowledge of the genomic profile of the ALK rearranged tumours at first diagnosis and monitoring for acquired resistance mechanisms using liquid biopsy approaches have potential to optimise ALK TKI and sequencing for improved outcomes.
Acknowledgments
Funding: This work is supported by International Association for the Study of Lung Cancer (IASLC) International Mentorship Award.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shaohua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.22). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 1994;263:1281-4. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol 2009;4:1450-4. [Crossref] [PubMed]
- Berge EM, Lu X, Maxson D, et al. Clinical benefit from pemetrexed before and after crizotinib exposure and from crizotinib before and after pemetrexed exposure in patients with anaplastic lymphoma kinase-positive non-small-cell lung cancer. Clin Lung Cancer 2013;14:636-43. [Crossref] [PubMed]
- U.S. Food & Drug Administration. Crizotinib. Available online: http://www.fda.gov/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CDER/ucm270058.htm
- U.S. Food & Drug Administration. FDA Approves Crizotinib. Available online: http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm376058.htm
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Marsilje TH, Pei W, Chen B, et al. Synthesis, structure-activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulfonyl)phenyl)pyrimidine-2,4-diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J Med Chem 2013;56:5675-90. [Crossref] [PubMed]
- Kinoshita K, Asoh K, Furuichi N, et al. Design and synthesis of a highly selective, orally active and potent anaplastic lymphoma kinase inhibitor (CH5424802). Bioorg Med Chem 2012;20:1271-80. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Kim DW, Mehra R, Tan DS, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 2016;17:452-63. [Crossref] [PubMed]
- Ou SH, Ahn JS, De Petris L, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol 2016;34:661-8. [Crossref] [PubMed]
- Nokihara H, Hida T, Kondo M, et al. Alectinib (ALC) versus crizotinib (CRZ) in ALK-inhibitor naive ALK-positive non-small cell lung cancer (ALK+ NSCLC): Primary results from the J-ALEX study. J Clin Oncol 2016;34:abstr 9008.
- Soria JC, Tan DS, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Katayama R, Sakashita T, Yanagitani N, et al. P-glycoprotein Mediates Ceritinib Resistance in Anaplastic Lymphoma Kinase-rearranged Non-small Cell Lung Cancer. EBioMedicine 2015;3:54-66. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Toyokawa G, Seto T. Updated Evidence on the Mechanisms of Resistance to ALK Inhibitors and Strategies to Overcome Such Resistance: Clinical and Preclinical Data. Oncol Res Treat 2015;38:291-8. [Crossref] [PubMed]
- Lovly CM, McDonald NT, Chen H, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 2014;20:1027-34. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Yoshida T, Oya Y, Tanaka K, et al. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383-9. [PubMed]
- Pérez-Callejo D, Romero A, Provencio M, et al. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res 2016;5:455-65. [Crossref] [PubMed]
- Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. [Crossref] [PubMed]
- Tan CL, Lim TH, Lim TKh, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [PubMed]
- He W, Xu D, Wang Z, et al. Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget 2016; [Epub ahead of print]. [PubMed]
- Mack P, Banks K, Riess J, et al. OA06.01 Clinical Utility of Circulating Tumor DNA (ctDNA) Analysis by Digital next Generation Sequencing of over 5,000 Advanced NSCLC Patients. J Thorac Oncol 2017;12:s263-4. [Crossref]
- Niu X, Chen Z, Chuai S, et al. MINI01.04: ROS1 Fusion Chinese Lung Adenocarcinoma Patients Treated with Crizotinib Detected Using Next-Generation Genotyping from ctDNA: Topic: Medical Oncology. J Thorac Oncol 2016;11:S258. [Crossref] [PubMed]
- Wang Z, Cheng G, Han X, et al. Application of Single-Molecule Amplification and Resequencing Technology for Broad Surveillance of Plasma Mutations in Patients with Advanced Lung Adenocarcinoma. J Mol Diagn 2017;19:169-81. [Crossref] [PubMed]
- Molina-Vila MA, Mayo-de-Las-Casas C, Giménez-Capitán A, et al. Liquid Biopsy in Non-Small Cell Lung Cancer. Front Med (Lausanne) 2016;3:69. [Crossref] [PubMed]
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [PubMed]
- Aguado C, Giménez-Capitán A, Karachaliou N, et al. Fusion gene and splice variant analyses in liquid biopsies of lung cancer patients. Transl Lung Cancer Res 2016;5:525-31. [Crossref] [PubMed]
- Brinkmann K, Emenegger J, Hurley J, et al. Exosomal RNA-based liquid biopsy detection of EML4-ALK in plasma from NSCLC patients. NCCN. Hollywood, 2016.