
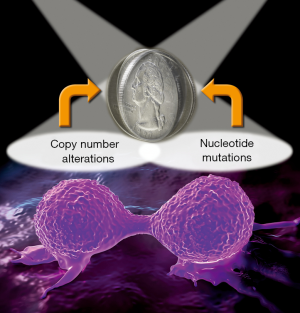
Haploinsufficiency and mutation are two sides of the cancer coin as cause and therapeutics target
Unprecedented intensity in the investigation of cancer genomes over the past 20 years has firmly established genome instability as a central hallmark of cancer. Publications from international consortia, particularly The Cancer Genome Atlas (TCGA), have painstakingly cataloged the genomic alterations that are frequently observed in all major cancer types (1-5). There are two primary categories of genome alterations: mutations at the nucleotide level and alterations at the level of chromosomal copy number (deletion and amplification). In some cancer types, such as colorectal cancer (CRC), genomic instability can be broadly classified as having chromosome instability (CIN) or microsatellite instability (MSI) (6,7). Whereas a majority of research investigation and drug development has been targeted on missense mutations (8,9), CIN has clearly been shown to play a key role in cancer development and progression and is associated with poor prognosis and drug resistance (10,11). Therefore, copy number alterations represent a wellspring for both research and therapeutics that have been underexplored. In this regard, nucleotide mutations and copy number alterations are really two sides of the same genomics instability coin for tumorigenesis. With the recent realization of the “randomness” in gene mutations due to replication errors in cancer (12), perhaps copy number alterations provide less randomness in therapeutic design.
In a recent publication in Nature Communications, Delaney and colleagues explored mathematical methodologies to map haploinsufficiency networks to identify targetable patterns of allelic deficiency (13). Ovarian cancer (OV) is the third most common gynecological cancer but ranks highest in mortality rate due to a high incidence of metastasis and development of resistance to frontline chemotherapies. Thus, there is a strong motivation to better understand the molecular underpinnings in the genomics of OV to develop more effective therapeutic strategies.
Delaney and colleagues analyzed data from OV patients of the TCGA cohort and found that most had low mutation rates in the well-known tumor driver genes, whereas levels of somatic copy-number-alterations (SCNAs) were high. They hypothesized that the SCNAs might play crucial roles in tumorigenesis and consequently impact clinical outcomes and indicate possible drug targets.
In this report, Delaney and colleagues proposed a novel network-based approach to identify significantly altered pathways consisting of gene-level disruptive SCNAs and to estimate the contribution of responsible genes within the altered pathways. They termed this innovative new analysis program the Haploinsufficient/Triplosensitive Gene (HAPTRIG) tool. An “edge score” was defined based on the GISTIC value and haploinsufficient annotation of each gene within the edge. They further proposed a “pathway score” to include the edge scores to prioritize pathways. Genes in the pathways were indexed by a “gene score”, which was defined as the sum of edge scores involving the node gene. Moreover, the HAPTRIG tool integrated information including gene-level SCNA, public databases about gene sensitivity to SCNA, and knowledge bases of protein interactions, to score disturbed pathways and contributed genes.
Using the HAPTRIG tool, the authors sought to infer impactful pathways and provide molecular mechanisms that bridged the SCNA landscape to the OV phenotypes. First, gene-level copy number alterations were derived from the segment-level SCNA landscape. Genes that were potentially sensitive to copy number deletion (haploinsufficient) or amplification (triplosensitive) were determined by mapping public databases of Yeast (YeastMine) and mouse (MouseMine) data to their human counterparts (Homology). Secondly, the biological impact of copy number alterations on SCNA-sensitive and insensitive genes were summarized as “pathway phenotypes” within 187 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Examples of possible pathway phenotypes included “reduced”, “severe” and “minimal/none”. Suppression of KEGG pathway activities due to copy number reduction of haploinsufficient genes were specifically prioritized, assuming copy number deletions were more deleterious than amplifications. These analyses led the authors to identify autophagy and proteostasis as the most disruptive pathways, and the BECN1 and LC3B as the most impactful genes in low mutation OV. They hypothesized that the copy number reduction of BECN1 and LC3B suppressed autophagy and disturbed proteostasis, which was responsible for poor outcomes and the drug resistance. To validate this hypothesis, BECN1 and LC3B were knocked down in a series of OV cell lines and in vivo models, including (I) OVCAR3, a cisplatin-resistant cell line that was genetically similar with high-grade OV patients; (II) IGROV1 and SKOV3, low SCNA OV cell lines; (III) OVCAR5 and OVCAR8; (IV) the patient-derived xenograft model cells LPPDOV and A2780. The experiments demonstrated that suppression of BECN1 and LC3B desensitized drug resistant high-grade OV cell lines to chemotherapies.
The authors also applied HAPTRIG to 20 other cancer types in the TCGA database and found variable levels of alterations in autophagy and proteostasis pathways. Furthermore, most proteostasis pathways in their pan-pathway analysis were enriched for deletions, including ER stress, ubiquitin-mediated proteolysis and the lysosome. The authors further suggested that the association of haploinsufficiency in model organism with an inability to form adequately proportioned protein-quality control complexes supported the notion that single allele SCNAs could disrupt these pathways. Notably, all CRC cases were analyzed as a whole in their analysis and results suggested modest suppression of proteostasis relative to high levels observed for OV, but it would be interesting to analyze the CIN and MSI subgroups of CRC separately. A recent publication has shown that haploinsufficiency in a 1p36 tumor suppressor gene, MIIP, in CRC plays a critical role in CRC metastasis (14). One of the recognized functions of MIIP is to regulate protein degradation/turnover mediated by the ubiquitin system (15,16). Therefore, a convergence of understanding in CIN appears to emerge from these independent studies.
It should be noted that Delaney and colleagues focused on copy number alterations. It can be envisioned, however, that the same algorithms could be used to explore the role of epigenetic alterations, which may functionally play the same role as haploinsufficiency. Interesting, the authors pointed out that KRAS mutant cancers were susceptible to elevated autophagy. Pancreatic adenocarcinoma (PAAD) is one of the prominent cancers with KRAS mutations, occurring in almost in all of the cases (17-19). PAAD is also known as a low mutation cancer type (20,21). In the report by Delaney and colleagues, PAAD did not show such highly suppressed autophagy as in OV cases based on the haploinsufficiency analysis. It would be intriguing to integrate data from methylome analysis. Relevant to this notion is the recent finding that methylation modulated expression of X-chromosome microRNA 506 (miR-506) is a potent inducer of autophagy-mediated cell death in PAAD (22).
Clearly, the recent Nature Communications paper by Delaney and colleagues has rightfully shed light on the often “ignored” side of the cancer coin, demonstrating the value of study of haploinsufficiency networks in genomic cancer instability (Figure 1). The HAPTRIG tool developed by the authors should lead more investigators to explore this wellspring of opportunities to benefit cancer prognosis and treatment.
Acknowledgments
Funding: W Zhang is supported by the endowed Hanes and Willis Professorship in Cancer and a Fellowship from National Foundation for Cancer Research. Y Sun is supported by the National Nature Science Foundation of China (No. 81472263 and 81201651), a grant from the Tianjin Municipal Science and Technology Commission (No. 14JCYBJC27500), and the National Key Clinical Specialist Construction Programs of China (No. 2013-544).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Zheng Li (Department of Gynecologic Oncology, The Third Affiliated Hospital of Kunming Medical University (Yunnan Tumor Hospital), Kunming, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.05.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Integrated genomic and molecular characterization of cervical cancer. Nature 2017;543:378-84. [Crossref] [PubMed]
- Grady WM. Genomic instability and colon cancer. Cancer Metastasis Rev 2004;23:11-27. [Crossref] [PubMed]
- Roepman P, Schlicker A, Tabernero J, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer 2014;134:552-62. [Crossref] [PubMed]
- Tie J, Gibbs P, Lipton L, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer 2011;128:2075-84. [Crossref] [PubMed]
- Bournet B, Buscail C, Muscari F, et al. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur J Cancer 2016;54:75-83. [Crossref] [PubMed]
- Nowak MA, Komarova NL, Sengupta A, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci U S A 2002;99:16226-31. [Crossref] [PubMed]
- Watanabe T, Kobunai T, Yamamoto Y, et al. Chromosomal Instability (CIN) Phenotype, CIN High or CIN Low, Predicts Survival for Colorectal Cancer. J Clin Oncol 2012;30:2256-64. [Crossref] [PubMed]
- Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 2017;355:1330-4. [Crossref] [PubMed]
- Delaney JR, Patel CB, Willis KM, et al. Haploinsufficiency networks identify targetable patterns of allelic deficiency in low mutation ovarian cancer. Nat Commun 2017;8:14423. [Crossref] [PubMed]
- Sun Y, Ji P, Chen T, et al. MIIP haploinsufficiency induces chromosomal instability and promotes tumour progression in colorectal cancer. J Pathol 2017;241:67-79. [Crossref] [PubMed]
- Ji P, Smith SM, Wang Y, et al. Inhibition of gliomagenesis and attenuation of mitotic transition by MIIP. Oncogene 2010;29:3501-8. [Crossref] [PubMed]
- Wen J, Fu J, Ling Y, et al. MIIP accelerates epidermal growth factor receptor protein turnover and attenuates proliferation in non-small cell lung cancer. Oncotarget 2016;7:9118-34. [PubMed]
- Bryant KL, Mancias JD, Kimmelman AC, et al. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci 2014;39:91-100. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Almoguera C, Shibata D, Forrester K, et al. Most Human Carcinomas of the Exocrine Pancreas Contain Mutant C-K-Ras Genes. Cell 1988;53:549-54. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Sun L, Hu L, Cogdell D, et al. MIR506 induces autophagy-related cell death in pancreatic cancer cells by targeting the STAT3 pathway. Autophagy 2017;13:703-14. [Crossref] [PubMed]


