
Multiplex screening of 422 candidate serum biomarkers in bladder cancer patients identifies syndecan-1 and macrophage colony-stimulating factor 1 as prognostic indicators
Introduction
Urothelial bladder cancer (UBC) is the 9th most common cancer world-wide (1). Approximately three-quarters of new cases present as non-muscle invasive disease (NMIBC) and the remainder present as muscle-invasive disease (MIBC). MIBC is life-threatening and patients require radical treatment with chemotherapy followed by cystectomy or radiotherapy (2). NMIBC patients are stratified into low-, intermediate- and high-risk groups according to clinicopathological parameters and are treated accordingly: transurethral resection (TUR) and intravesical mitomycin C for low risk, and TUR and intravesical BCG for intermediate- and high-risk NMIBC (3). Radical cystectomy is also considered an option for high risk NMIBC. UBC is initially detected by flexible cystoscopy, and NMIBC patients are subjected to regular long-term surveillance with this burdensome procedure. Urine or serum biomarkers that could detect and characterise UBC non-invasively could improve management of bladder cancer patients in several ways, e.g.,
- Detecting primary tumours—to facilitate the triage of haematuria patients into those at high or low risk of having UBC;
- Detecting recurrent tumours—to reduce the reliance on cystoscopy for NMIBC surveillance;
- Prognosis—to improve existing clinicopathological prognostication and select more appropriate treatment regimens for individual patients;
- Prediction—to predict which patients will benefit from which therapeutic agents;
- Staging—to determine which patients have MIBC at presentation and move directly to cross-sectional imaging and definitive radical therapies.
Considerable effort has been expended on identifying urinary biomarkers for detecting the presence of bladder cancer with many urine markers proposed, ranging from FDA-approved assays (e.g., NMP22, BTA) to numerous markers reported in single studies (4). Most of the proposed markers are proteins, and none retain both high (clinically useful) sensitivity and specificity upon large scale validation. More recently, DNA-based urine biomarkers have shown considerable promise and have re-awakened the hope that non-invasive disease detection with clinically useful sensitivity and specificity may be achievable (5-8).
Most prognostic studies have measured biomarkers directly on tumour tissue, although there are reports of biomarker levels in urine and plasma/serum providing prognostic information. Changes in DNA methylation, gene expression profiles and individual protein levels have shown promise as prognostic indicators in research studies [reviewed in (9)]. As with detection, no prognostic biomarkers have yet been widely accepted into clinical practice. In addition to detection and prognostic roles, predictive biomarkers are likely to find a place in the management of bladder cancer as new targeted therapeutic agents are adopted.
A biomarker test that assisted in non-invasive staging of bladder cancer could facilitate the fast-tracking of MIBC patients to cross-sectional imaging and definitive treatment, circumventing delays and possible disease dissemination due to TUR. An experienced urologist can distinguish between low-grade papillary NMIBC and high-grade (HG) solid tumours. However, discrimination between HG T1 NMIBC and MIBC is difficult to achieve at cystoscopy (10) and hence staging TUR is undertaken—an invasive and possibly detrimental procedure that could be avoided if there was an alternative for detecting muscle-invasion. Whilst a pinch biopsy of tumour may be collected during cystoscopy for biomarker analysis, for HG tumours mutation and gene expression profiles appear to traverse stages (11): a discernible “molecular switch” which enables (and hence indicates) muscle invasion has not yet been identified. Levels of molecules released directly from tumours or by tissue degradation into body fluids during invasion might better identify disease stage. Levels of urinary biomarkers are typically higher in MIBC then NMIBC patients; however, the NMIBC subgroup is comprised of low-grade and pTa disease in addition to HG T1 disease. The link between grade and biomarker concentration usually appears much stronger than the link between stage and biomarker concentration, and hence discrimination between HG T1 and MIBC disease based on urinary biomarkers is challenging. We hypothesised that non-invasive detection of MIBC might more effectively be achieved using serum/plasma biomarkers. Recently, levels of both circulating tumour cells and plasma ctDNA have shown promise as staging and prognostic markers (12-14). Nonetheless, the extremely low levels of these biomarkers in the circulation make them challenging to measure, whereas proteins are easier and faster to measure, potentially enabling point-of care testing. Additionally, only the “tip-of–the-iceberg” of the plasma proteome has been explored in bladder cancer patients to date, leaving plenty of potential for biomarker discovery. A literature search identified just over 20 proteins that have been investigated as serum markers for bladder cancer and that show a stage dependent increase in concentration. However, none substantially increase between HG NMIBC and MIBC; the largest increase is typically seen in stages T3 and T4 [e.g., MMP7 and HNPs1-3 (15,16)], most likely reflecting the largest increase in tumour burden. Indeed, levels of some of these serum biomarkers are highly prognostic and can predict non-organ-confined disease (17), but there are no convincing reports of serum biomarkers distinguishing between NMIBC and MIBC.
We reasoned that since none of the protein biomarkers reported to date looks highly promising for non-invasive staging, then future analyses should be broader to include more proteins. Mass spectrometry-based “shotgun” proteomics is the method of choice for identifying and quantitating large numbers of proteins in biological and clinical specimens (18). This approach is limited in terms of sample throughput, but more so in ability to detect low abundance proteins in serum: potentially relevant cytokines and tumour leakage products may be 109 fold more dilute than abundant serum proteins and so may not be detectable. Conversely, individual protein assays such as ELISAs would be prohibitively expensive and time-consuming if a large number of proteins were to be considered. We therefore utilised a novel multiplex assay which enables simultaneous measurement of 92 proteins in 90 samples using “proximity extension” and requiring only 1 µL of sample. In this method, a pair of oligonucleotide-conjugated antibodies is used for each analyte. When both antibodies bind to an analyte molecule the close proximity of the oligonucleotides enables ligation, extension and amplicon generation. The amplicons are subsequently analysed by qPCR (http://www.olink.com/) giving relative quantitation of all analytes across all samples.
Methods
Patient samples
All sera were collected as part of the Bladder Cancer Prognosis Programme between 2004 and 2011 (UK ethics ref: 06/MRE04/65). Full details of this multi-centre biospecimen collection have been published elsewhere (19). Briefly, patients whose diagnostic cystoscopy indicated primary bladder cancer were recruited to the study, and blood collected into serum tubes prior to TUR. The blood was left to clot for 90–150 min, centrifuged at 3,500 rpm for 10 min and the sera stored at −80 °C. Ultimately, some of the patients were diagnosed with non-malignant conditions and these serve as non-cancer controls. All patients were followed for at least 3 years following initial diagnosis. Patient information is summarised in Table 1. Age and gender did not differ significantly between the patient groups.
Full table
Assays
Protein concentrations in patient sera were analysed using five Proseek Multiplex panels (Oncology, Inflammation, Neurology, Cardiovascular II and III) at the Proseek Multiplex Analysis Laboratory (http://www.olink.com/). Each Proseek assay measures 92 proteins. Briefly, a pair of oligonucleotide-conjugated antibodies to each protein are added to 1 µL of serum. When an antibody-protein-antibody sandwich is formed both antibodies are in close proximity, the oligonucleotides hybridize, and an extension reaction forms a unique sequence. These sequences are then quantitated by microfluidic qPCR.
Data analysis
All data were analysed as normalised protein expression (NPX on a log2 scale). The patients were separated into five classes of increasing stage and grade: non-UBC, G1pTa, G3pTa, G3T1 and G3T2+. Associations between serum protein concentrations and stage/grade groups were tested using multivariate linear regression analysis with age and gender as covariates using R statistical software version 3.2.5. Multiple testing corrections were done with the Benjamini-Hochberg method and an adjusted P<0.05 was considered significant. Significant proteins were further investigated using ROC analyses and t-tests to compare NMIBC and MIBC patients and Kaplan-Meier analyses and log rank testing to study survival using SigmaPlot 12.5. For survival analyses the 80 UBC patients were divided into “high” and “low” according to whether their biomarker concentration was above or below the median value for the UBC patients (control subjects excluded).
Results
Assay performance
Sera from 90 patients were analysed on five Proseek multiplex immunoassay panels (92 analytes each). Patient characteristics are shown in Table 1. Quality control criteria for the datasets were considered good with 97–99% of the samples meeting QC criteria across the five Proseek panels and 94% of proteins being above the limit of detection in 100% of the samples. Due to redundancy between panels and some analytes not meeting QC criteria, reportable results were obtained for 422 unique proteins. In instances of proteins being measured on multiple panels good agreement between data from the panels was observed (data not shown). A list of all 422 analytes is provided in Supplemental Information.
Proteins associated with UBC
The concentrations of proteins in the sera of the non-UBC, G1pTa, G3pTa, G3pT1 and G3pT2+ UBC patients were compared using multivariate linear regression. Whilst 80 proteins appeared to be significantly associated with bladder cancer (P<0.05), this was reduced to five following Benjamini-Hochberg multiple testing correction (Supplemental Information). The five proteins were (in order of statistical significance, lowest P value first) nectin-4, syndecan-1, T-cell immunoglobulin mucin receptor 1, macrophage colony-stimulating factor 1 and MMP7. Boxplots of the serum concentrations of these proteins in the five patient groups are shown in Figure 1.

Protein associations with UBC stage
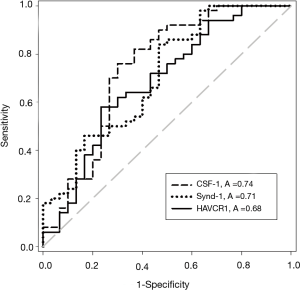
The concentrations of all five UBC associated proteins show a positive correlation with disease stage and all five of the UBC associated proteins were significantly more concentrated in the sera of patients with MIBC than NMIBC (P<0.05, t-test). Table 2 shows the relative concentrations of these proteins in NMIBC (n=50) and MIBC patients (n=30). Despite being significantly elevated in MIBC patients relative to NMIBC patients, the concentration changes are modest and the areas under the ROC curves below 0.75. The distributions of the three most discriminatory proteins in the NMIBC and MIBC patients are shown in Figure 2 and corresponding ROC curves are shown in Figure 3. The heat-map shown in Figure 4 shows the distribution and heterogeneity of the 5 significant proteins’ concentrations in the patient sera. Approximately half of the MIBC cases have elevated levels of several or all of these proteins, but there are also MIBC cases where none of the biomarkers are elevated. Thus, whilst high-levels of these proteins are highly indicative of MIBC, it would not be possible to devise a sensitive test for MIBC using any combination of these proteins.

Full table


Protein associations with outcome
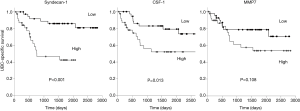
We investigated the relationship between the levels of the five UBC associated proteins and bladder cancer specific survival. With all five proteins there was a trend towards poorer outcome with high biomarker levels; this approached statistical significance in the case of MMP7 and was highly significant in the cases of syndecan-1 and CSF-1 (Figure 5). Syndecan-1 does not reach significance if we only consider NMIBC patients (P=0.081, 50 patients, 7 UBC-specific deaths) but is highly significant in the MIBC patient group (P<0.001, 30 patients, 16 UBC-specific deaths).
Discussion
We have used multiplex immunoassays to measure the concentrations of 422 proteins in the serum of bladder cancer patients. The relationship between the serum concentrations of many of these proteins and UBC have not been previously reported. Disappointingly, no accurate biomarkers for non-invasively detecting or staging bladder cancer have been uncovered. Nonetheless, the serum concentrations of five of the proteins investigated are statistically significantly associated with bladder cancer and two of the proteins show an association with reduced bladder cancer specific survival. Several of the UBC associated proteins are trans-membrane proteins which are most likely present as soluble forms in the serum due to ectodomain shedding (20). The elevated serum concentrations may be due to increased expression within the tumour, increased sheddase activity in the tumour microenvironment, or a combination of both. Consistent with the current findings, we have previously reported that increased urinary concentrations of the shed ectodomains of EpCAM and EGFR indicate a poor prognosis (20,21)
The most significantly bladder cancer-associated protein was nectin-4, a calcium-independent transmembrane cell-adhesion molecule which has not previously been investigated in UBC, and is expressed both in normal urothelium and HG UBC at moderate to high-levels (The Human Protein Atlas). Nectin-4 did not discriminate well between NMIBC and MIBC or show significant prognostic potential but has previously been reported as a biomarker for breast, lung and ovarian cancers (22-24), and it is believed that the extracellular domain is shed into the circulation via ADAM17-mediated cleavage (25).
Syndecan-1 was the second most cancer-associated protein in multivariate analysis, and also shows some discrimination between NMIBC and MIBC. Like nectin-4, syndecan-1 is a transmembrane protein most likely shed into the circulation by ADAM17-mediated cleavage (26). We find that elevated serum syndecan-1 is a very significant indicator of poor outcome in UBC patients. This is in agreement with the study by Szarvas et al. in which serum syndecan-1 was found to be an independent prognostic indicator in a cohort of 79 patients (27).
Our third UBC-associated molecule, T-cell immunoglobulin mucin receptor 1 (HAVCR1), is again a transmembrane protein whose extracellular domain can be released into the circulation by proteolytic cleavage (28). There are no publications relating to serum levels of this protein in UBC patients, but it is reportedly overexpressed in renal and ovarian cancers and an anti-HAVCR1 antibody-drug conjugate is currently being developed to treat renal, lung and ovarian cancers (29).
Macrophage colony stimulating factor 1 (CSF-1) is our fourth most cancer-associated protein and shows the best discrimination between NMIBC and MIBC of any of the proteins studied. It is a secreted cytokine (released by ectodomain shedding) which is released by several UBC cell lines (30); increased serum concentrations have been reported in several cancer types including breast, lung and ovarian (31-33) and are associated with a poor prognosis. Higher CSF-1 serum levels in some UBC cases may reflect high levels of tumour associated macrophages, which itself is associated with worse prognosis (34).
Matrix metalloprotease 7 (matrilysin or MMP7) is a secreted protease involved in tissue remodelling which has previously been detected at increased levels in advanced bladder cancer; it has been reported to be an independent prognostic indicator in a study of 79 UBC patients (16). Whilst our results corroborate the finding of elevated serum MMP7 in advanced UBC, serum MMP7 failed to reach statistical significance as a prognostic indicator in our study.
Conclusions
We have studied the relationship between UBC and the serum concentrations of 422 proteins. Those which reach statistical significance have been discussed and include both known biomarkers and new candidates that may warrant further investigation. Although UBC does cause many biologically plausible changes in the serum proteome, the disease is highly heterogeneous and none of the proteins studied appears to be suitable for non-invasive staging of bladder cancer. Our data suggest that the majority of the proteins studied here do not merit further investigation as UBC serum biomarkers, with the possible exceptions of syndecan-1 and CSF-1 which do appear to be highly prognostic. A major strength of our work is the use of the Proseek platform which has enabled, for the first time, measurement of hundreds of low abundance proteins in the sera of bladder cancer patients. The limitations of our study include the modest sample size and that the Proseek assays are a biomarker discovery tool that provides relative quantitation of a large number of proteins across a patient cohort rather than being an approved clinical test. To develop the latter would require either developing a custom “bladder cancer panel” multiplex assay (possibly using Proseek technology) or running one or more ELISAs. Validation in independent prospective studies is required to determine if these proteins are robust prognostic indicators and to determine whether they provide information over and above clinicopathological parameters.
Acknowledgments
We would like to thank Emil Nilsson (OLINK proteomics) for his help with data analysis.
Funding: This work was funded by a donation to the University of Birmingham for bladder cancer research.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Ja Hyeon Ku, Kunyoo Shin, Minyong Kang) for the series “Bladder Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.04.19). The series “Bladder Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by NRES Committee East Midlands—Derby (UK ethics ref: 06/ MRE04/65) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Alfred Witjes J, Lebret T, Compérat E, et al. Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer. Eur Urol 2017;71:462-75. [Crossref] [PubMed]
- Babjuk M, Böhle A, Burger M, et al. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur Urol 2017;71:447-61. [Crossref] [PubMed]
- D'Costa JJ, Goldsmith J, Wilson J, et al. A Systematic Review of the Diagnostic and Prognostic Value of Urinary Protein Biomarkers in Urothelial Bladder Cancer. Bladder Cancer 2016;2:301-17. [Crossref] [PubMed]
- Reinert T, Modin C, Castano F, et al. Comprehensive genome methylation analysis in bladder cancer: identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin Cancer Res 2011;17:5582-92. [Crossref] [PubMed]
- Tognieri FS, Ward DG, Foster JM, et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet 2016;24:1167-74. [Crossref] [PubMed]
- van Kessel KE, Beukers W, Lurkin I, et al. Validation of a DNA methylation-mutation urine assay to select patients with hematuria for cystoscopy. J Urol 2017;197:590-5. [Crossref] [PubMed]
- Ward DG, Baxter L, Gordon N, et al. Multiplex PCR and next generation sequencing for the non-invasive detection of bladder cancer. PLoS One 2016;11:e0149756 [Crossref] [PubMed]
- van Rhijn BW, Catto JW, Goebell PJ, et al. Molecular markers for urothelial bladder cancer prognosis: toward implementation in clinical practice. Urol Oncol 2014;32:1078-87. [Crossref] [PubMed]
- During VA, Sole GM, Jha AK, et al. Prediction of histological stage based on cytoscopic appearances of newly diagnosed bladder tumours. Ann R Coll Surg Engl 2016;98:547-51. [Crossref] [PubMed]
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15:25-41. [Crossref] [PubMed]
- Christensen E, Birkenkamp-Demtröder K, Nordentoft I, et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur Urol 2017;71:961-9. [Crossref] [PubMed]
- Gazzaniga P, de Berardinis E, Raimondi C, et al. Circulating tumor cells detection has independent prognostic impact in high-risk non-muscle invasive bladder cancer. Int J Cancer 2014;135:1978-82. [Crossref] [PubMed]
- Soave A, Riethdorf S, Dahlem R, et al. A nonrandomized, prospective, clinical study on the impact of circulating tumor cells on outcomes of urothelial carcinoma of the bladder patients treated with radical cystectomy with or without adjuvant chemotherapy. Int J Cancer 2017;140:381-9. [Crossref] [PubMed]
- Gunes M, Gecit I, Pirincci N, et al. Plasma human neutrophil proteins-1, -2, and -3 levels in patients with bladder cancer. J Cancer Res Clin Oncol 2013;139:195-9. [Crossref] [PubMed]
- Szarvas T, Becker M, vom Dorp F, et al. Matrix metalloproteinase-7 as a marker of metastasis and predictor of poor survival in bladder cancer. Cancer Sci 2010;101:1300-8. [Crossref] [PubMed]
- Margel D, Bostrom P, Baniel J, et al. External validation of a biomarker based pre-cystectomy algorithm to predict nonorgan confined urothelial cancers. J Urol 2012;187:840-4. [Crossref] [PubMed]
- Csősz É, Kalló G, Márkus B, et al. Quantitative body fluid proteomics in medicine - A focus on minimal invasiveness. J Proteomics 2017;153:30-43. [Crossref] [PubMed]
- Zeegers MP, Bryan RT, Langford C, et al. The West Midlands Bladder Cancer Prognosis Programme: rationale and design. BJU Int 2010;105:784-8. [Crossref] [PubMed]
- Bryan RT, Shimwell NJ, Wei W, et al. Urinary EpCAM in urothelial bladder cancer patients: characterisation and evaluation of biomarker potential. Br J Cancer 2014;110:679-85. [Crossref] [PubMed]
- Bryan RT, Regan HL, Pirrie SJ, et al. Protein shedding in urothelial bladder cancer: prognostic implications of soluble urinary EGFR and EpCAM. Br J Cancer 2015;112:1052-8. [Crossref] [PubMed]
- Derycke MS, Pambuccian SE, Gilks CB, et al. Nectin 4 overexpression in ovarian cancer tissues and serum: potential role as a serum biomarker. Am J Clin Pathol ;134:835-45. [Crossref] [PubMed]
- Fabre-Lafay S, Monville F, Garrido-Urbani S, et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer 2007;7:73. [Crossref] [PubMed]
- Takano A, Ishikawa N, Nishino R, et al. Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer Res 2009;69:6694-703. [Crossref] [PubMed]
- Fabre-Lafay S, Garrido-Urbani S, Reymond N, et al. Nectin-4, a new serological breast cancer marker, is a substrate for tumor necrosis factor-alpha-converting enzyme (TACE)/ADAM-17. J Biol Chem 2005;280:19543-50. [Crossref] [PubMed]
- Pruessmeyer J, Martin C, Hess F, et al. A disintegrin and metalloproteinase 17 (ADAM17) mediates inflammation-induced shedding of syndecan-1 and -4 by lung epithelial cells. J Biol Chem 2010;285:555-64. [Crossref] [PubMed]
- Szarvas T, Reis H, Kramer G, et al. Enhanced stromal syndecan-1 expression is an independent risk factor for poor survival in bladder cancer. Hum Pathol 2014;45:674-82. [Crossref] [PubMed]
- Schweigert O, Dewitz C, Möller-Hackbarth K, et al. Soluble T cell immunoglobulin and mucin domain (TIM)-1 and -4 generated by A Disintegrin And Metalloprotease (ADAM)-10 and -17 bind to phosphatidylserine. Biochim Biophys Acta 2014;1843:275-87.
- Thomas LJ, Vitale L, O'Neill T, et al. Development of a Novel Antibody-drug Conjugate for the Potential Treatment of Ovarian, Lung and Renal Cell Carcinoma Expressing TIM-1. Mol Cancer Ther 2016;15:2946-54. [Crossref] [PubMed]
- Steube KG, Meyer C, Drexler HG. Secretion of functional hematopoietic growth factors by human carcinoma cell lines. Int J Cancer 1998;78:120-4. [Crossref] [PubMed]
- Scholl SM, Lidereau R, de la Rochefordière A, et al. Circulating levels of the macrophage colony stimulating factor CSF-1 in primary and metastatic breast cancer patients. A pilot study. Breast Cancer Res Treat ;39:275-83. [Crossref] [PubMed]
- Będkowska GE, Ławicki S, Gacuta E, et al. M-CSF in a new biomarker panel with HE4 and CA 125 in the diagnostics of epithelial ovarian cancer patients. J Ovarian Res 2015;8:27. [Crossref] [PubMed]
- Kaminska J, Kowalska M, Kotowicz B, et al. Pretreatment serum levels of cytokines and cytokine receptors in patients with non-small cell lung cancer, and correlations with clinicopathological features and prognosis. M-CSF - an independent prognostic factor. Oncology 2006;70:115-25. [Crossref] [PubMed]
- Hanada T, Nakagawa M, Emoto A, et al. Prognostic value of tumor-associated macrophage count in human bladder cancer. Int J Urol 2000;7:263-9. [Crossref] [PubMed]


