
The up-regulation of TANK-binding kinase 1 in head and neck squamous cell carcinoma
IntroductionOther Section
Head and neck squamous cell carcinoma (HNSCC), which accounts for more than 90 percent of head and neck malignant tumors, is the sixth most common cancer type worldwide (1,2). Every year, there were about 600,000 new cases diagnosed as HNSCC and approximately in men 2 to 3 times higher than in women around the world (3,4). In clinical practice, we have noticed that one of the most important causes of poor prognosis is lymph node metastasis, which often had been occurred prior to the diagnosis. What’s more, metastasis could decrease about half of the survival rate of HNSCC (5). Hence, investigating the related proteomics of HNSCC and searching for a biomarker is very meaningful.
TANK-binding kinase 1 (TBK1) is also named as NF-kB activating kinase (NAK), and it shares about 65 percent similarity to IKKɛ and belongs to a non-canonical IkB kinase (6,7). TBK1 has 729 amino acids and approximately stable expression in all types of human tissues (8,9). Recently some studies have revealed that the high expression of TBK1 protein was related to the occurrence and development of human malignant tumors (10-12).
Therefore, we designed the study and tried to investigate the function of TBK1 in HNSCC. We first analyzed the GEO datasets and then verified the results using immunohistochemistry and siRNA transfection, and the goal of the present study was to clarify whether TBK1 could be offer additional help for the early diagnosis or even targeted therapy of HNSCC.
MethodsOther Section
Cell culture and main reagents
The HNSCC cell lines, FaDu and Cal-27, were cultured in RMPI1640 (Invitrogen) with 10% fetal bovine serum (FBS) at 37 °C, 5% CO2. FBS was obtained from HyClone (Logan, UT, USA). RPMI-1640 media and 0.25% trypsin solution were purchased from Invitrogen (Carlsbad, CA, USA). Cells in logarithmic growth phase were used for western blotting.
Gene expression profiles
GEO dataset GSE59102 (HOX genes: potential candidates furthering the development of larynx squamous cell carcinoma), GSE51985 (microarray gene expression analysis of tumorigenesis and regional lymph node metastasis in laryngeal squamous cell carcinoma), GSE58911 (gene expression in normal and tumor samples from patients with HNSCC), GSE10774 (HNSCC cell lines compared with normal keratinocytes), GSE27020 (identification and validation of a multigene predictor of recurrence in primary laryngeal cancer), was considered to investigate TBK1 expression level in HNSCC tumor tissue and normal tissue, and to investigate the correlation between TBK1 expression level and tumor differentiation and prognosis.
Patients selected and tissue microarray (TMA)
An independent TMA (Xi’an Alenabio company, China) (Production number: LP481) was purchased. The TMA consisted of 30 tumor tissues and 8 normal larynx or pharynx tissues, including 31 men and 7 women. The mean age was 51.2 years old (ranged from 15 to 72 years old) (Table 1).
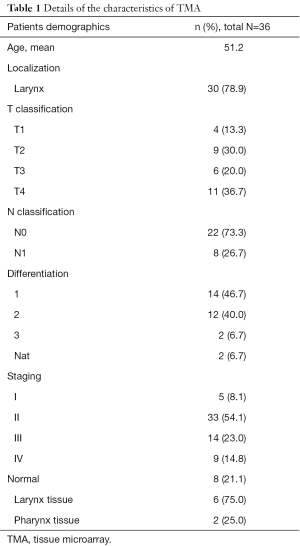
Full table
Immunohistochemical staining
The TMA was deparaffined with standard pure xylene for 15 minutes three times at room temperature and hydrated in graded alcohols, phosphate buffer saline (PBS) was used to wash the TMA. Antigen retrieval was performed in boiling citrate buffer (PH 6.0) for 15 minutes. Then TMA was cooled down to room temperature in the buffers. After washing TMA in PBS for 5 minutes three times, 0.3% hydrogen peroxide phosphate-citrate buffer was used to block endogenous peroxidase activity for 10 minutes. Rinsed with PBS for 5minutes, TMA was incubated with primary antibody TBK1 (Sigma-Aldrich; dilution 1:100) for 12 hours at 4 °C. The TMA was incubated with Poly-HRP Goat anti-rabbit (Maixin. Bio, FuZhou, China) for 30 minutes. Slides were dyed with diaminobenzidine for 5 minutes. Haematoxylin was used to counterstain the nucleus, followed by dehydration and mounted. Images of TMA were taken using an Olympus BX40 microscope and CC-12 Soft-Imaging System (Olympus, Tokyo, Japan).
Evaluation of immunohistochemical staining
The TMA was analyzed and scored for intensity [0–3] and frequency [0–4]. The intensity was scored as grade [0], negative; grade [1], weak intensity; grade [2], moderate intensity; grade [3], strong intensity. The frequency scores were respectively assigned when 0–25%, 26‒50%, 51–75% and 76–100% of the tumor cell were positive. To use statistical analysis, TBK1 protein intensity and frequency were transformed into a Composit Expression Score (CES) utilizing the formula CES = Intensity × Frequency. The range of CES was from 0 to 12. The CES was scored as negative [0], weak positive [1–4], positive [5–8], strong positive [9–12].
Synthesis of siTBK1 gene and gene transfection
The siRNAs molecules used for suppression of FaDu and Cal-27 cell lines TBK1 gene and the negative control siRNA (which does not target any sequence present in the cell genome) were all obtained from RiboBio Co Ltd (Guangzhou, China). Transient transfection of siRNA was carried out with the lipofectamine 2000 regent (Invitrogen, Massachusetts, USA) according to the manufacturer’s instructions.
Cell proliferation
Cells were seeded at 1×103/well in 96-well plates in RPMI-1640 medium supplemented with 10% FBS. Twenty-four hours later, the cells were transfected, and proliferation was determined at 0, 24, 48 and 72 h post-transfection using the Cell Counting Kit-8 (CCK-8), according to the manufacturer’s instructions.
Flow cytometry for detection of cell apoptosis
Cells were harvested by trypsinization and washed twice with cold PBS. Then the cells were resuspended in Annexin-V-binding buffer and then stained with 5 µL of Annexin-VFITC solution and 10 µL of propidium-iodide (PI) solution for 15 min in dark at room temperature. Fluorescence was analyzed on a FACSCantoTM II spectrometer (BD Biosciences, San Jose, CA, USA). Cells stained with FITC/PI were counted as apoptotic cells.
Western blotting
Total cell lysate was performed according to standard instructions. The lysates were resolved using 10% SDS-PAGE, transferred to PVDF membranes and immunoblotted with primary antibodies against TBK1, GAPDH, p-TBK1, T-TBK1, p-NF-kB p65, T-NF-kB p65, livin and survivin. Following incubation with secondary antibodies, the protein bands were detected using an enhanced chemiluminescence reagent (Thermo Fisher Scientific, Rockford, IL, USA).
Statistic analysis
All statistical analyses were performed using the Statistical Package for Social Sciences (SPSS) version 20.0 software. The big data downloaded from GEO database was imported into excel table for further statistical analysis. The differences between the expressions of TBK1 in tumor and normal tissues were compared by using t-test, and the differences among the expressions of TBK1 in tumor tissues with different degree of differentiation were analyzed by using chi-square test. P value <0.05 was considered statistically significant.
ResultsOther Section
The significantly higher expression of TBK1 in HNSCC tissue
GEO datasets GSE59102, GSE51985, GSE58911, GSE10774 and GSE27020, were analyzed. We compared the relative expression of TBK1 mRNA in the normal tissue and HNSCC in four GEO datasets (GSE59102, GSE51985, GSE58911, GSE10774). It revealed that TBK1 expression in tumor was higher than normal tissue (Figure 1A, ID: A_23_P44768, P=0.0059; Figure 1B, ID: ILMN_1739967, P=0.0028; Figure 1C, ID: 7956795, P=0.0294; Figure 1D, ID: 8134, P=0.0138). These data indicated that TBK1 gene expression level in HNSCC was up-regulated.
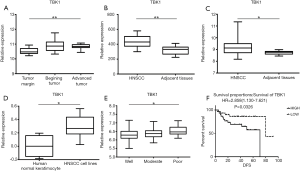
The significantly higher expression of TBK1 in poorly differentiated tumor
After that, we investigated the correlation between TBK1 expression level and tumor differentiation in GSE27020. It showed that TBK1 expression in poorly differentiated tumor was significantly higher (Figure 1E, ID: 218520_at, P=0.0375).
The poor prognosis of the patients with higher expression of TBK1
We also investigated the correlation between TBK1 gene expression and prognosis of HNSCC patients. The result showed that patients with higher expression of TBK1 had poorer prognosis [Figure 1F, ID: 207324_s_at, HR=2.858 (1.130–7.621), P=0.0326], suggesting that the different expression level of TBK1 may be correlated with the prognosis of the patients with HNSCC.
Analysis of TBK1 expression in TMA
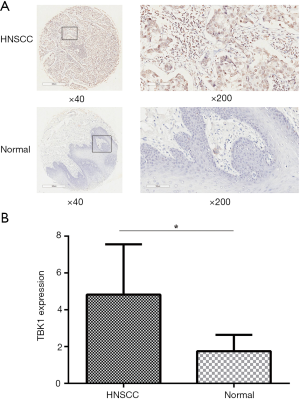
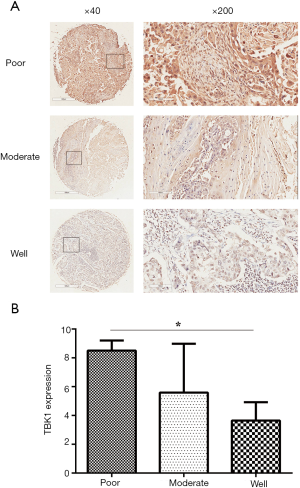
In order to completing an analysis of TBK1 in clinic samples of HNSCC, we used TMA to verify the results obtained from GEO datasets. The CES scores of every sample of the TMA were measurement. We chose the images of TBK1 expression from the TMA, representative images were shown in Figure 2A. TBK1 expression in HNSCC was significantly higher than normal tissue (Figure 2B, P<0.05), and it was also higher in poorly differentiated tumor tissues than well differentiated tumor tissues (Figure 3, P<0.05). The result was also consistent with what we analyzed from the GEO datasets.
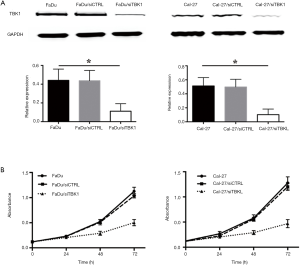
TBK1 suppressing in FaDu and Cal-27
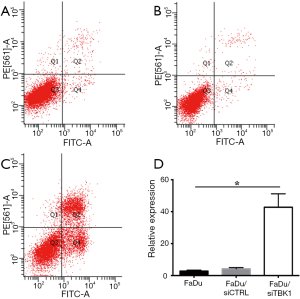
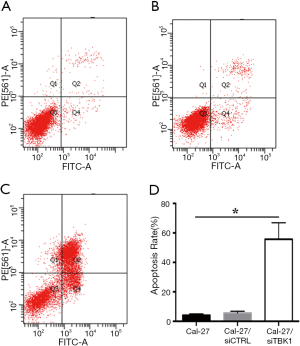
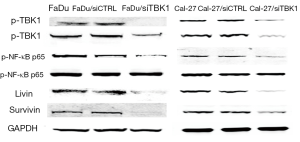
FaDu and Cal-27 cells transfected with TBK1 siRNA (named as FaDu/siTBK1 and Cal-27/siTBK1). Control cells were transfected with nonsense siRNA (named as FaDu/siCTRL and Cal-27/siCTRL). Western blot revealed that the expression of TBK1 protein was significantly decreased in FaDu/siTBK1 and Cal-27/siTBK1 cells (Figure 4A, P<0.05). After TBK1 suppressing the proliferation rate of FaDu and Cal-27 cells were inhibited in a time-dependent manner compared with the control cells (Figure 4B, P<0.05). Moreover, the proportion of apoptotic cells of FaDu/siTBK1 (Figure 5, P<0.05) and Cal-27/siTBK1 (Figure 6, P<0.05) was significantly higher than control cells. Finally, the results of Western blot also showed that several relevant proteins such as livin and survivin were down-regulated in FaDu/siTBK1 and Cal-27/siTBK1 (Figure 7).
DiscussionOther Section
HNSCC is the most important type of head and neck cancer and the anatomical location of it is very complex (13), the main treatment methods are surgical operation, chemotherapy and radiotherapy. Nowadays although some markers, such as p63, CK5/6 and so on, have been found or even applied on clinic (14,15), the application and effectiveness of them are still not very satisfactory. Because of this, more studies should be done to investigate the genomics and proteomics of HNSCC.
In recent years, a number of studies have revealed that TBK1 was related to various types of human malignant tumors, particularly lung cancer and breast carcinoma (16,17). And in most cancer cells TBK1 always showed higher protein expression than normal (18,19), which was in accordance with the result observed in the present study.
We analyzed the TBK1 mRNA expression in GEO datasets and found that TBK1 mRNA was up-regulation in HNSCC, and then the result had been verified by TMA test. In addition, the result of GSE27020 showed that TBK1 expression level was also related to tumor differentiation and prognosis of HNSCC.
Wei et al. (20) investigated the expression of TBK1 in breast cancer and suggested that high TBK1 expression showed high potential for relapse. However, in another paper the researchers inhibited TBK1 expression and revealed the loss of TBK1 expression could help to promote lung metastasis in breast cancer (21). Hence, the effect of TBK1 on prognosis of HNSCC still need further study.
Furthermore, in order to clarify the cellular mechanism of TBK1 in HNSCC we silenced TBK1 gene and found that the proportion of apoptotic cancel cells increased significantly. The same phenomenon was observed in the researches about TBK1 in lung cancer.
When TBK1 was inhibited in A549 human lung cancer cells, the epithelial–mesenchymal transition (EMT) would be attenuated via activation of GSK-3β and repression of ZEB1 (22). In another study about lung cancer, the investigator indicated that TBK1 was a mitosis regulator and knockdown of TBK1 could decrease the metadherin phosphorylation, and then promote the growth inhibition and apoptosis of cancer cells (23). Both of the two mechanisms might also exist in HNSCC.
However, Muvaffak et al. pointed out that TBK1 might be a therapeutic target but the proliferation and apoptosis were not solely dependent on TBK1 (7). Some other genes such as miR-203 also could suppress tumor growth by targeting TBK1 in osteosarcoma (24). In addition, livin and survivin proteins are both belong to IAP protein family, which is an inhibitor apoptosis protein family. The suppression of TBK1 can result in the down-regulation of livin and survivin proteins expression showed that TBK1 might also be related to IAP protein family.
In conclusion, the present study revealed that TBK1 gene expression was higher in HNSCC than normal tissue, and it might offer additional help for the early diagnosis. TBK1 was correlated to tumor differentiation and prognosis of HNSCC, the inner mechanism was not very clear. What’s more, TBK1 might be a meaningful therapeutic target of HNSCC and mechanisms need further investigation.
AcknowledgmentsOther Section
Funding: This work was supported by the National Natural Science Foundation of China (No. 81372880, 81172569); the doctoral program of Higher Education Research Fund (No. 20130141120093, 20110141110062); and the Natural Science Foundation of Hubei province (No. 2012FFA045).
FootnoteOther Section
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Zhou Y, Peng Y, Tang H, et al. Autophagy induction contributes to GDC-0349 resistance in head and neck squamous cell carcinoma (HNSCC) cells. Biochem Biophys Res Commun 2016;477:174-80. [Crossref] [PubMed]
- Khammanivong A, Sorenson BS, Ross KF, et al. Involvement of calprotectin (S100A8/A9) in molecular pathways associated with HNSCC. Oncotarget 2016;7:14029-47. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Busch CJ, Becker B, Kriegs M, et al. Similar cisplatin sensitivity of HPV-positive and -negative HNSCC cell lines. Oncotarget 2016;7:35832-42. [Crossref] [PubMed]
- Mamelle G, Pampurik J, Luboinski B, et al. Lymph node prognostic factors in head and neck squamous cell carcinomas. Am J Surg 1994;168:494-8. [Crossref] [PubMed]
- Shen RR, Hahn WC. Emerging roles for the non-canonical IKKs in cancer. Oncogene 2011;30:631-41. [Crossref] [PubMed]
- Muvaffak A, Pan Q, Yan HY, et al. Evaluating TBK1 as a therapeutic target in cancers with activated IRF3. Mol Cancer Res 2014;12:1055-66. [Crossref] [PubMed]
- Larabi A, Devos JM, Ng SL, et al. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep 2013;3:734-46. [Crossref] [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Kim JY, Welsh EA, Oguz U, et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci U S A 2013;110:12414-9. [Crossref] [PubMed]
- Tu D, Zhu Z, Zhou A, et al. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep 2013;3:747-58. [Crossref] [PubMed]
- Kim JY, Beg AA, Haura EB. Non-canonical IKKs, IKKϵ and TBK1, as novel therapeutic targets in the treatment of non-small cell lung cancer. Expert Opin Ther Targets 2013;17:1109-12. [Crossref] [PubMed]
- Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer 2010;46:765-81. [Crossref] [PubMed]
- Wang J, Zhang X, He J, et al. Co-expression of EGFR and CK5/6 in primary squamous cell carcinoma of the breast. Med Oncol 2014;31:172. [Crossref] [PubMed]
- Ma Y, Fan M, Dai L, et al. Expression of p63 and CK5/6 in early-stage lung squamous cell carcinoma is not only an early diagnostic indicator but also correlates with a good prognosis. Thorac Cancer 2015;6:288-95. [Crossref] [PubMed]
- Newman AC, Scholefield CL, Kemp AJ, et al. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS One 2012;7:e50672 [Crossref] [PubMed]
- Deng T, Liu JC, Chung PE, et al. shRNA kinome screen identifies TBK1 as a therapeutic target for HER2+ breast cancer. Cancer Res 2014;74:2119-30. [Crossref] [PubMed]
- Kim JK, Jung Y, Wang J, et al. TBK1 regulates prostate cancer dormancy through mTOR inhibition. Neoplasia 2013;15:1064-74. [Crossref] [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Wei C, Cao Y, Yang X, et al. Elevated expression of TANK-binding kinase 1 enhances tamoxifen resistance in breast cancer. Proc Natl Acad Sci U S A 2014;111:E601-10. [Crossref] [PubMed]
- Yang KM, Jung Y, Lee JM, et al. Loss of TBK1 induces epithelial-mesenchymal transition in the breast cancer cells by ERα downregulation. Cancer Res 2013;73:6679-89. [Crossref] [PubMed]
- Liu W, Huang YJ, Liu C, et al. Inhibition of TBK1 attenuates radiation-induced epithelial-mesenchymal transition of A549 human lung cancer cells via activation of GSK-3 beta and repression of ZEB1. Lab Invest 2014;94:362-70. [Crossref] [PubMed]
- Kim JY, Welsh EA, Oguz U, et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci U S A 2013;110:12414-9. [Crossref] [PubMed]
- Liu S, Feng P. MiR-203 determines poor outcome and suppresses tumor growth by targeting TBK1 in osteosarcoma. Cell Physiol Biochem 2015;37:1956-66. [Crossref] [PubMed]


