Circulating tumor DNA detection in advanced non-small cell lung cancer patients
Introduction
Non-small cell lung cancer (NSCLC) accounts for 85–90% of lung cancers (1). It has been the most common malignancy and the leading cause of cancer death for decades. Currently, the 5-year overall survival for NSCLC is still lower than 15% worldwide (2). Thus precise detection of gene mutations at an early stage is of great benefit to improve patient treatment. Although the tumor tissue biopsies are the gold standard for mutation detection, some inherent shortcomings still exist in clinical practice, such as the tumor heterogeneity (3,4). Therefore, novel detection methods are urgently needed to characterize circulating tumor DNA (ctDNA) mutations.
ctDNA is a class of DNA fragments that originate from tumor cells; it can be extracted from blood plasma (5,6). Due to its sources, ctDNA carries tumor-specific mutations from tumor cells (7). The detection of ctDNA mutations can reflect the overall genetic profiles of tumors, which may avoid the tumor heterogeneity problems in tissue biopsies. Moreover, the ctDNA samples can be obtained easily and repeatedly taken over the process of patient treatment, and thus can be applied to real-time and dynamical monitoring of evolving gene mutations to guide patient treatment (8,9). Therefore, ctDNA is of great value in reflecting the gene mutations of NSCLC patients, thereby highlighting the potential of ctDNA detection in personalized medicine.
In recent years, various methods have been developed to detect ctDNA mutations at different scales, such as PCR-based and NGS-based methods (10-13). PCR-based methods can accurately detect ultra-low frequency mutations, but few gene mutations can be detected simultaneously (14). Because cancer development involves multiple gene alterations, it is difficult to comprehensively characterize cancer genetic information with PCR-based methods. The NGS-based methods mainly involve two methods, such as the amplicon-based method and the hybrid capture method (12,13). The high cost of the probes, high background noise, and the longtime of hybridization time limit the application of the hybrid capture method in clinical practice (15). High background noise limits the application of the amplicon-based method. Recently, the amplicon-based method has been combined with molecular barcoding to reduce the background errors (14). This method is called tag sequencing. However, evaluations of its performance are lacing, so its performance needs to be further validated.
In the present study, we evaluated the performance of tag sequencing. We first detected mutations in a reference standard cfDNA set with four concentrations (0.0%, 0.1%, 1.0% and 5.0%) to evaluate its performance. Moreover, we analyzed the mutation concordance between plasma DNA and matched tissue DNA by comparing the epidermal growth factor receptor (EGFR) mutations in 20 Chinese advanced NSCLC patients.
Methods
Patient features
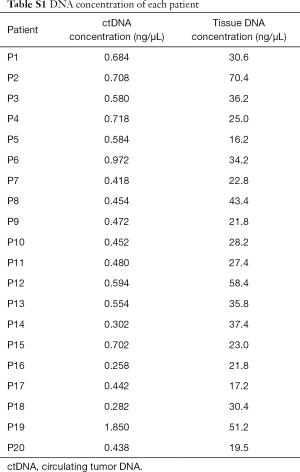
Twenty NSCLC patients were recruited from Zhujiang Hospital, Southern Medical University, Guangdong Province, China. The Ethical and Protocol Review Committee of Zhujiang Hospital approved the study protocol. Written informed consent was obtained from all patients. Table S1 lists the detailed clinical information and demographic features of patients.
DNA preparation
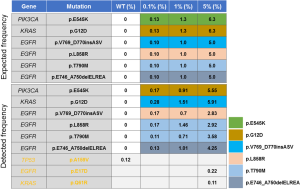
For the reference standard cfDNA, a multiplex I cfDNA Reference Standard Set was obtained from Horizon (Horizon Discovery, Cambridge, United Kingdom) with four different concentrations (0.0%, 0.1%, 1.0%, and 5.0%). The multiplex I cfDNA Reference Standard Set included six known mutations, which could be detected by our assay. The allele frequency of each mutation was validated by droplet digital PCR, as shown in Figure 1. For the 20 NSCLC patients, plasma and formalin-fixed paraffin embedded (FFPE) tissue samples were examined for mutation profiles. Genomic DNA was extracted from sections of FFPE tissue samples using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Venous blood samples were collected in K2-EDTA tubes. Then the tubes were spun twice, first at 1,800 g for 10 minutes and then at 16,000 g for 15 minutes. The plasma-depleted whole blood was stored at −80 °C for cfDNA isolation. ctDNA was extracted from blood using the MagMAX cfDNA extraction kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. From each phlebotomy specimen, 1.5–4 mL (median 2.5 mL) of plasma was profiled to target ~20 ng of cfDNA input into Tag-Seq library preparation (Table S1).
NGS library construction
The Oncomine™ Lung cfDNA Kit (Life Technologies) was applied to construct the adapter-ligated library. Library quality control was performed using the Qubit®2.0 and 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). A multiplexed of 16 libraries consisting of 100 pM prepared library per sample, which were then amplified by emulsion PCR on Ion Sphere™ Particles (ISPs) with the IonOneTouch™ 2 Instrument (Life Technologies). Finally, the template-positive ISPs were enriched and loaded onto and run on Ion Proton (Life Technologies). A panel covering more than 150 hotspot mutations in 11 cancer-related genes, such as EGFR, ALK, BRAF, KRAS, PIK3CA, and TP53, was used in this study.
Mutation identification
The adaptor sequences of the raw data were removed, and then the clean reads were mapped to the human reference genome (hg19). The hotspot and targeted region, together with the parameter files associated with the Oncomine™ Lung cfDNA Assay, were loaded into the variantCaller plugin (Life Technologies) to call and to filter the mutations. The mutations were further filtered using the following filters: (I) the minimum allele frequencies ≥0.1%; (II) the minimum coverage of the mutations ≥1,000×. All identified mutations were visually confirmed by using the Integrative Genomics Viewer (IGV) (16).
Amplification refractory mutation system (ARMS)-PCR validation
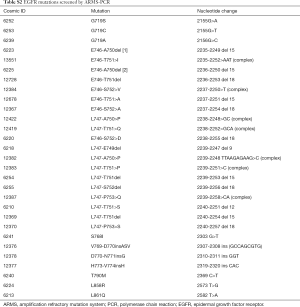
To validate the mutations detected by tag sequencing in the plasma ctDNA, the matched tissues of 20 NSCLC patients were screened for 29 known EGFR mutations (Table S2) using ARMS-PCR as previously described (17).
Results
Patient features
Tissues and matched blood samples were obtained from 20 NSCLC patients, including 7 females and 13 males. Table 1 lists the patient clinical characteristics. Participants in this study cohort were diagnosed with stage IIIB to IV NSCLC, and all had adenocarcinoma (100%). The majority of patients (12/20, 60%) were non-smokers and most of them (18/20, 90%) were treatment naive. In the two treatment-experienced patients, one had received first-line target therapy for the past 1.5 years and the other was undergoing chemotherapy.

Full table
NGS data coverage analysis
All samples, consisting of 20 ctDNA samples from NSCLC patients and 4 different concentrations of reference standard cfDNA, were sequenced using Ion Proton. The mean depth of NSCLC patient samples was 335,801, and the range was from 111,359 to 489,020. The mean depth of the reference standard cfDNA samples was 122,991, with a range from 52,994 to 178,002.
NGS assay performance evaluated using the cfDNA reference standard set
Evaluation of assay performance, including accuracy and stability, is our research priority. Therefore, we assessed assay performance by sequencing the cfDNA Reference Standard Set with four different concentrations (0.0%, 0.1%, 1.0%, and 5.0%) and then comparing their detected frequencies with their expected frequencies. In the 0.1%, 1.0%, and 5.0% groups, the assay could detect all six mutations and their allele frequencies were close to their expected frequencies (Figure 1). The true positive rate of these three groups was 100% and the false positive rate were 0%. In the 0.0% group, none of these six mutations were detected by the assay, which was concordant with their expected frequencies (0%) (Figure 1). In this group, the true negative rate was 100% and the false negative rate was 0%. In addition to these six mutations, one more TP53 mutation was detected in the 0.0% group and two more mutations were detected in the 5.0% group, which may be inherent mutations of the engineering cell line (Figure 1).
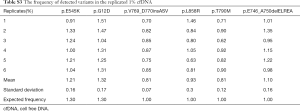
To further evaluate the assay stability, five more repeated experiments were sequenced in the 1% concentration group. In these six repeated experiments, the mean frequencies of four EGFR mutations (p.T790M, p.L858R, p.V769_D770insASV and p.E746_A750delELREA) were 0.81%, 0.93%, 0.81% and 1.1%, respectively (Table S3). The frequencies of these four mutations were all close to their expected frequencies 1% (Table S3). The mean frequencies of PIKCA3 (p.E545K) and KRAS (p.G12D) were 1.21% and 1.32%, respectively. The frequencies of these two mutations were all close to their expected frequencies 1.3% (Table S3). The standard deviation (SD) of these six mutations were all less than 0.18%, which may indicate the favorable stability of our assay (Table S3).
EGFR mutation concordance in matched tissue DNA and plasma ctDNA
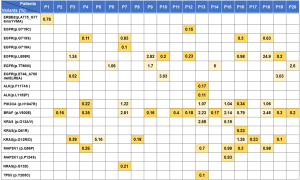
By using our assay, single-nucleotide variants (SNVs) and insertion-deletion polymorphisms (Indels) of the NSCLC-related genes, including EGFR, ALK, PIK3CA, BRAF, KRAS, NRAS, TP53, MAP2K1 and ERBB2, were detected in the plasma ctDNA samples of 20 NSCLC patients. Figure 2 presents the detected mutations for each patient and their frequencies.
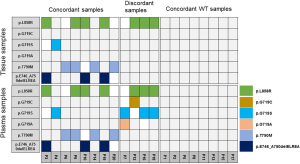
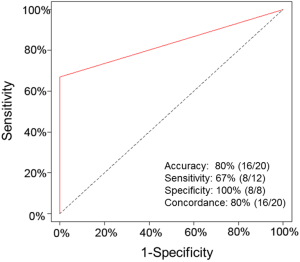
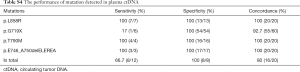
Because one of our major objectives was to compare the detection concordance between the matched tissue DNA and plasma ctDNA, we compared the EGFR mutations in the plasma and matched tissue samples of 20 NSCLC patients. In the plasma ctDNA, six EGFR mutations were detected: p.T790M, p.L858R, p.E746_A750delELREA, p.G719C, p.G719S, and p.G719A (Figure 3). Of the 12 EGFRm+ patients, all were concordant for the common sensitive mutations p.L858R, p.T790M, and p.E746_A750delELREA, however, the plasma test appears to result in false positive G719X calls in 4 cases. The 8 EGFRwt patients were all concordant in that no mutations were detected in tissue or plasma samples. The sensitivity and specificity has been shown in Table S4. According to patients, the sensitivity and specificity of detection mutations in plasma ctDNA and tissue DNA were 67% and 100%, respectively (Figure 4). The accuracy and the concordance were 80% and 80%, respectively (Figure 4). In summary, the concordant results between tag sequencing and the routine clinical approaches demonstrated that the tag sequencing method using the ctDNA has great potential in detecting gene mutations in NSCLC patients.
Discussion
In the plasma and the matched tissue samples obtained from 20 NSCLC patients, tag sequencing was applied to simultaneously detect multiple NSCLC-related gene mutations. Apart from the NSCLC samples, four different concentrations of a reference standard cfDNA set were used to evaluate the assay performance. For the NSCLC samples, the EGFR mutation concordance between the plasma ctDNA and tissue DNA was 80% and the sensitivity and specificity were 67% and 100%, respectively (Figure 4). For the reference standard cfDNA samples, the assay detected 100% of the six known mutations, and their detection frequencies were all close to the expected allele frequencies (Figure 1). The assay was very stable and the SDs of all mutations were less than 0.18% in the six replicated experiments (Table S3). These results indicate NGS has the great potential in detecting the gene mutations of tumors, so tag sequencing could be a promising tool in NSCLC diagnosis and personalized medicine.
The genetic profiles characterized by ctDNA can monitor tumor genetic alterations to guide patient treatment. Clinical testing for EGFR mutations is now a routine in case of NSCLC to guide treatment. NSCLC patients with p.L858R mutations tend to be sensitive to EGFR-TKIs (18,19),whereas the patients with the secondary p.T790M mutation tend to develop drug resistance to the first generation of EGFR-TKIs (20,21). In our study, both of these mutations could be detected in the plasma ctDNA (Figure 3). Patients P9 with both mutations, had developed resistance to treatment with EGFR-TKIs, such as gefitinib and erlotinib (Figure 4). For the patient P3, P10, P12, P16, P18 and P19, the plasma ctDNA could be applied to monitor whether the patients had developed the drug resistance (p.T790M) to EGFR-TKIs. Taken together, these findings indicate that the identification of ctDNA mutations has great benefits for tumor diagnosis, patient treatment and survival prediction.
Various methods have been developed to detect the mutations in the plasma ctDNA, such as PCR-based methods and NGS-based methods. The sensitivity of EGFR mutations detected in the ctDNA is 66.7% (34/51) by PNA-PCR (22), 72.1% (44/61) by ARMS (23) and 70.6% (12/17) by the NGS based on the hybrid capture method (15). In our study, we applied tag sequencing based on Ion Torrent to detect mutations in ctDNA. The sensitivity of EGFR mutations was approximately 70%, which was similar to other methods, but our NGS-based method can simultaneously detect a large number of gene mutations compared with PCR-based methods. Moreover, compared with the NGS-based hybrid capture method, our method is much less expensive and time consuming, making the technique more suitable for clinical use.
Although we found that tag sequencing accurately detected the mutations in the reference standard cfDNA set, and we observed high levels of concordance between plasma ctDNA and the matched tissue DNA, tour study had two major limitations. First, we assessed concordance merely using the EGFR mutations in 20 matched tissue and plasma samples. In future research, more genes, such as BRAF, KRAS, NRAS, PIK3CA, MAP2K1 and TP53, should be included to comprehensively investigate the concordance between plasma ctDNA and matched tissue DNA. Also, a larger sample size will be required to further assess the performance of our concordant analysis. Third, more reference mutations were required to assess the performance of our assay. In this study, we evaluated the performance of tag sequencing using the four concentrations of the reference standard cfDNA set containing six detected mutations. Although detection was successful with four reference standard cfDNA concentrations and six replicated experiments at 0.1%, more mutations should be included to evaluate assay performance in the future.
In summary, the tag sequencing method can effectively and stably detect gene mutations in four concentrations of a reference standard cfDNA set. Moreover, our results revealed the high concordance rates between plasma ctDNA and matched tissue DNA in 20 advanced NSCLC patients, suggesting that the ctDNA in plasma has great potential in characterizing the genetic profiles of tumors, so its application may be of great value in molecular diagnostics, prognosis prediction and targeted drug selection.
Conclusions
Our findings indicated that tag sequencing can effectively and stably detect gene mutations in ctDNA, and is suitable for clinical application. Tag sequencing accurately detects the ctDNA mutations, so its application may be of great value in molecular diagnostics, prognosis prediction and targeted drug selection.
Full table
Full table
Full table
Full table
Acknowledgments
Funding: This study was supported by Science and Technology Program of Guangdong (2015B020233009, 2015A030401040), China Postdoctoral Science Foundation funded project (2016M602486).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Ethical and Protocol Review Committee of Zhujiang Hospital. All procedures performed in studies involving human participants were in accordance with the ethical standards of Zhujiang hospital and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [Crossref] [PubMed]
- Albain KS, Crowley JJ, Turrisi AT 3rd, et al. Concurrent cisplatin, etoposide, and chest radiotherapy in pathologic stage IIIB non-small-cell lung cancer: a Southwest Oncology Group phase II study, SWOG 9019. J Clin Oncol 2002;20:3454-60. [Crossref] [PubMed]
- Tan DS, Camilleri-Broët S, Tan EH, et al. Intertumor heterogeneity of non-small-cell lung carcinomas revealed by multiplexed mutation profiling and integrative genomics. Int J Cancer 2014;135:1092-100. [Crossref] [PubMed]
- Majem M, Remon J. Tumor heterogeneity: evolution through space and time in EGFR mutant non small cell lung cancer patients. Transl Lung Cancer Res 2013;2:226-37. [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. [Crossref] [PubMed]
- Stroun M, Maurice P, Vasioukhin V, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci 2000;906:161-8. [Crossref] [PubMed]
- Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999;96:9236-41. [Crossref] [PubMed]
- Dressman D, Yan H, Traverso G, et al. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A 2003;100:8817-22. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [Crossref] [PubMed]
- Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Yao Y, Liu J, Li L, et al. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget 2017;8:2130-40. [PubMed]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178-92. [Crossref] [PubMed]
- Cai W, Lin D, Wu C, et al. Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J Clin Oncol 2015;33:3701-9. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Del Re M, Tiseo M, Bordi P, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget 2017;8:13611-9. [PubMed]
- Han JY, Choi JJ, Kim JY, et al. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer 2016;16:627. [Crossref] [PubMed]
- Chai X, Ren P, Wei B, et al. A comparative study of EGFR oncogenic mutations in matching tissue and plasma samples from patients with advanced non-small cell lung carcinoma. Clin Chim Acta 2016;457:106-11. [Crossref] [PubMed]