LncRNA TUSC7 affects malignant tumor prognosis by regulating protein ubiquitination: a genome-wide analysis from 10,237 pan-cancer patients
Introduction
Cancer is the uncontrolled growth of tissue caused by various carcinogenic factors. Worldwide, cancer is the leading cause of morbidity and mortality, with about 14 million new cases in 2012.While the overall cancer mortality in the USA has declined by 25%, over the past 20 years more than 2 million Americans have died of cancer (1). Effective tumor markers assist clinicians in detecting, understanding the pathogenesis and predicting the prognosis of cancer. Emerging researches have sought to discover more effective tumor markers from the perspective of non-coding RNA (2,3), but these remain far from clinical practice.
Long non-coding RNA (lncRNA) is a genre of non-coding transcripts with a length of over 200 nt, which regulates gene expression at various biological processes, including transcriptional and post-transcriptional regulation and other epigenetic regulation. Aberrant expression of lncRNA, as oncogenic or tumor suppressor genes, is in relation to the development of cancer. Some lncRNAs have been studied in translational research, such as clinical diagnostic, prognostic evaluation (4) and treatment response prediction (5,6), but further prospective clinical validation of these reports is required.
LncRNA tumor suppressor candidate 7 (TUSC7), also known as LOC285194 and antisense RNA LSAMP3, is located at 3q13.31, and has a length of 14,347 bases. TUSC7 is expressed at low levels in tumor samples, relative to the control groups, in lung cancer, esophageal cancer, gastric cancer, liver cancer and other tumors (7-11), suggesting that it may be involved in cancer development. However, how TUSC7 epigenetically regulates downstream proteins remains unclear.
In the present translational analysis, we initially conducted meta- analyses to examine the relationship of TUSC7 expression and the clinical-pathological factors of patients with solid malignancies. The association with survival outcome was further analyzed. In addition, the Cancer Genome Atlas (TCGA) data sets were used to validate the results of the literature analysis. A broad-spectrum signal pathway analysis exhibited that TUSC7 potentially regulates the ubiquitination process. In summary, our findings imply that TUSC7 is differentially expressed in cancer and is associated with prognosis in cancer patients by its impact on protein ubiquitination.
Methods
Study selection and data extraction
The China National Knowledge Infrastructure (CNKI), Google Scholar, PubMed and Science Direct databases were searched for reports published prior to June 2017, using the key words “long non-coding RNA”, “LSAMP3”, “TUSC7”, “LOC285194” and “cancer” to find eligible studies. The included studies met the following criteria: (I) the TUSC7 expression levels in both primary tumor and paracancerous/normal tissue was examined; (II) included cancer patients with clinical data; and (III) sufficient survival data to calculate the hazard ratio (HR), odds ratios (ORs) the and 95% confidence intervals (CI). In articles presenting survival curve without HR and CI, specific points from the survival curves were extracted by Engauge Digitizer version 4.1 and the survival data were obtained according to Tierney et al. (12). If there were duplicate data sets, the most complete or the most recent data was chosen. Exclusion criteria were as follows: (I) there were no available or insufficient data; (II) case reports; and (III) meeting abstracts. Two researchers independently extracted and reviewed relevant data from the studies, including the first author, publication year, country, cancer site, methods, number of cases, and cut-off values. If a decision could not be unanimously agreed upon, a third researcher intervened to reach a consensus.
Study quality and publication bias assessment
Literature retrieval investigators (Yusong Chen and Xiaoshun Shi) assessed the quality of all studies by reading the titles and abstracts independently and the Newcastle-Ottawa quality assessment scale was applied to evaluate the quality of literature. In the present meta-analysis, publication bias of the incorporated studies was assessed and illustrated by funnel plots. If the estimated point by the combined effect after the deletion of a study fell below the 95% CI of combined effect value, it manifested that the study had a significant impact on the combined effects.
Pan-cancer RNA-seq data and survival data from TCGA
Publicly available Level 3 RNA-seq data (HTSeq-FPKM-UQ) and the corresponding clinical data of 10,237 cancer patients were obtained from TCGA data portal website (http://cancergenome.nih.gov). The expression levels of TUSC7 and prognosis data were extracted. The HR and 95% CI were subjected to the Survival Analysis R package (version 3.0) for further meta-analysis.
Acquisition of microarray data
The 60,000 sets of microarray data were obtained from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). The gene expression data were retrieved from the EBI ArrayExpress database by Bioconductor (13,14) and pre-processed using the Robust Multichip Average (RMA) normalization method. Datasets with low standard deviation levels were filtered out, and the remaining experimental data were used for the co-expression analysis.
Gene set enrichment analysis (GSEA)
Lung adenocarcinoma (LUAD) gene expression profiles were accessed and downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/). The GSEA was applied to mine the relevant biological processes. Briefly, the top and bottom quartiles of TUSC7 expression (high and low TUSC7 expression, respectively) were sorted, then default settings were used and the gene sets with FDR of 0.25 was regarded as cutoff for the identification of biologically relevant genes. The enriched pathways in each phenotype were sorted by the normalized enrichment score (NES) and nominal P value.
Statistical analysis
The R (version 3.0) statistical package was used to extracted survival data from the TCGA data, STATA software version 12 (Stata, Corporation, College, Station, Texas, USA) was utilized for meta-analysis and evaluation of heterogeneity among studies was performed using the Cochrane Q test and P values. If there was heterogeneity (I2 ≥50% or P≤0.05), the pooled OR was calculated using the random effect model. Otherwise, the fixed effect model is applied. All P values were two tailed, and the sensitivity analysis and publication bias were considered statistically significant if P<0.05.
Results
Characteristics of included studies
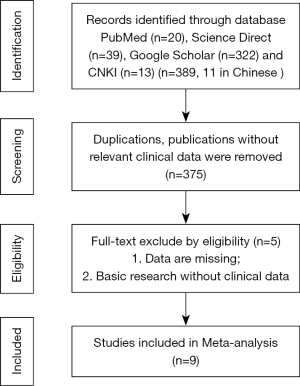
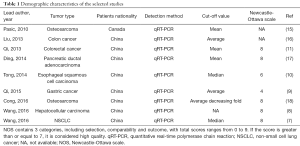
The initial database search identified 389 articles published between 2010 and 2016 (Figure 1). After screening the titles and abstracts, 14 articles were further evaluated for eligibility. Of these, 5 papers were excluded because they contained either no data or incomplete data sets. Thus, 9 articles were included in the present meta-analysis. Eight different types of cancer were analyzed, including 1 study each of esophageal carcinoma, gastric cancer, hepatocellular carcinoma, non-small cell lung cancer, pancreatic ductal carcinoma and 2 studies each regarding colorectal cancer and osteosarcoma. These studies enrolled 655 participants, with a minimum sample size of 75 and a maximum sample size of 142, and the levels of TUSC7 expression in both normal or paracancerous tissues and tumor tissues were determined. The main features of the 9 studies and the Newcastle-Ottawa scale (NOS) score are summarized in Table 1. Since TUSC7 expression was detected by qPCR, the cut-off values differed between these studies. Not all studies reported the association between TUSC7 expression and sex, tumor size, differentiation, lymph node metastasis, distant organ metastasis and TNM staging or other indicators.
Full table
Association between TUSC7 and clinicopathological characteristics of cancers
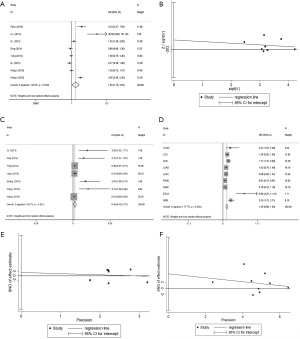
As shown in Figure 2A, the differential expression of TUSC7 in cancer and paracancerous tissues and its association with clinicopathological features of patients were meta-analyzed. The results displayed that TUSC7 is expressed differentially between cancerous and paracancerous tissues (HR =1.833, 95% CI: 1.102–3.048). Our statistic outcome suggests that TUSC7 is differentially expressed in the existing cancer studies and sensitivity analysis showed no publication bias (P=0.567) in these studies (P>0.05) (Figure 2B). Unfortunately, the outcomes demonstrated that the expression levels of TUSC7 showed no associations with age, gender, tumor differentiation, tumor size, lymph node metastasis or TNM staging (Figure S1).
TUSC7 is associated with cancer survival
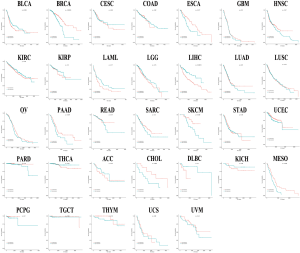
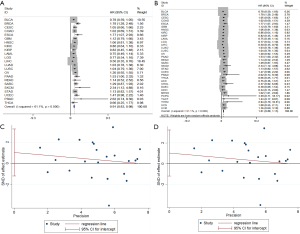
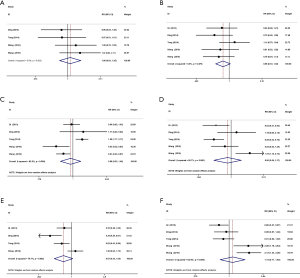
To investigate further the effect of differential expression of TUSC7 in cancer, 7 studies with OS data from 616 patients were chosen for meta-analysis (Figure 2C). We applied a random effects model due to heterogeneity (I2 =56.7%, P=0.03). Low TUSC7 expression was in association to a poorer survival rates, and TUSC7 expression had a protective role in cancer prognosis (HR =0.436, 95% CI: 0.099–0.773, P=0.02). According to the type of malignancy included in our studies, corresponding survival data of these tumors were portrayed (Figure S2) and extracted from the TCGA database for survival analysis (Table S1). With the results which from survival analysis were subjected to meta-analysis (Figure 2D). The pooled survival data corresponding to the literature in the TCGA database displayed that low TUSC7 expression is associated with a poorer survival rate and is related to an adverse prognosis in cancer (HR =1.25, 95% CI: 0.88–1.63, P=0.00). This result implied that TUSC7 has impact on cancer survival in existing cancer studies. No clear evidence of publication bias (P=0.83 and P=0.38, respectively) in this meta-analysis were able to find (Figure 2E,F).
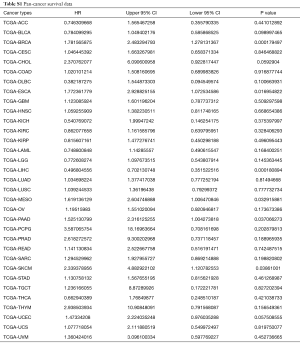
TUSC7 is a prognostic determinant of pan-cancer survival
TCGA program has published the genetic data for 33 cancers. To explore the broad relationship between TUSC7 and clinical outcomes, we divided the patients into high and low expression groups according to the median value of TUSC7 expression, and analyzed survival data using R (Table S1, Figure S2). After meta-analysis of the clinical and genetic data from 10,237 samples in the TCGA dataset, we confirmed that low TUSC7 expression is associated with poorer survival and is also associated with adverse outcomes in both common cancer types (HR =1.019, 95% CI: 0.88–1.16, P=0.00) (Figure 3A) and a pan-cancer scope (HR =1.01, 95% CI: 0.88–1.13, P=0.00) (Figure 3B). There was no clear evidence of publication bias (P=0.34 and P=0.51, respectively) in these meta-analyses (Figure 3C,D).
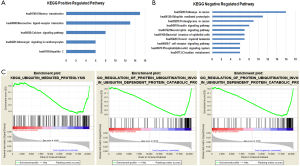
The association of TUSC7 with ubiquitin mediated proteolysis pathway
Next, we aimed to reveal potential mechanisms to explain the effects of TUSC7 on cancer prognosis. The co-expression RNA transcripts of TUSC7 were identified by analysis of its co-expression genes from over 60,000 public Affymetrix human U133-Plus 2 transcriptional profiling microarray data, including all normal and cancer tissues. The top five positive or ten negative co-expressed genes significantly enriched pathways were showed (Figure 4A,B). Bioinformatics analysis externally validated that TUSC7 is downregulated in most cancer tissues, compared to normal or paracancerous tissues, and gene sets are significantly enriched in the low TUSC7 phenotype. To identify further key pathways associated with cancer prognosis, LUAD RNAseq2 data from the TCGA were subjected to the GSEA assay. We discover that ubiquitin mediated proteolysis pathway related genes were significantly enriched in the LUAD patients with low TUSC7 expression (Figure 4C). Taken together, our results indicate that TUSC7 plays a prognostic role in cancer patients by regulating ubiquitination.
Discussion
In the study of the human genome, it has been reported that most genome sequences are transcribed into non-coding RNAs (19). LncRNA is a class of non-coding RNA, having a length of more than 200 nt, which has been considered as a non-functioning genomic transcription. However, recent studies advised that lncRNAs have impacts in many cancers (20), such as lung cancer (21), breast cancer (22), and prostate cancer (23). At present, abnormal expression of lncRNAs have been found in a variety of tumor tissues, which plays a therapeutic role in the treatment of human malignant tumors (24). These findings suggest that lncRNAs have potential roles in cancer translational medicine.
LncRNA TUSC7 was first identified as a tumor suppressor unit in osteosarcoma (15). Studies have reported that the expression of TUSC7 in cancer tissues is lower than normal tissues. However, the role of TUSC7 in cancer biology is far from fully elucidated. Pasic et al. (15) reported that TUSC7 is a transcriptional target of p53 and that ectopic expression of TUSC7 inhibits colorectal and breast cells growth. Deletion analysis showed that two miR-211 binding sites are associated with TUSC7-mediated growth inhibition in colorectal carcinoma (16). However, the mechanism by which TUSC7 affects gene expression modification and protein interaction has not been studied. In our findings, TUSC7 is down-regulated on the pan-cancer level and is associated with cancer prognosis. By screening TUSC7 relevant genes and mining potential molecular pathways, we first reported that TUSC7 is widely involved in the process of protein ubiquitination.
Post-transcriptional ubiquitination can affect chromatin function and plays an important regulatory role in gene expression. Yoon et al. found that lncRNA HOTAIR could induce ubiquitin mediated proteolysis (25). The increased expression of HOTAIR could promote the growth and invasion of prostate cancer cells. It was found that lncRNA HOTAIR combined with androgen receptor in cancer cells, blocking the binding of E3 ubiquitin ligase MDM2 and AR, thereby preventing the ubiquitination and promoting the degradation of AR (26). Moreover, lncRNA-mediated ubiquitination contributes to the biological process of protein stability. For example, lncRNA ANCR can promote the interaction of CDK1-EZH2, increasing the phosphorylation of Thr-345 and Thr-487 in EZH2, and facilitating the ubiquitination process to degrade the targeted protein (27). By genome-wide and extensive tissue gene co-expression analysis, TUSC7 has been considered to be involved in the process of protein ubiquitination. Further biological pathway mining from the lung cancer TCGA RNA-seq data provided an independent, external validation.
However, our analysis has some limitations regarding the correlation of aberrant expression of lncRNA and clinical significance. First, the definition of low or high levels of TUSC7 was different in terms of cut-off values and detection methodology. Although real-time quantitative PCR was applied to quantify TUSC7 in all studies, the results could still have been heterogeneous due to different qPCR primer designs. Second, the follow-up schedule and differing endpoints of the TCGA studies limit the accuracy of the results. Third, most of the patients in the literature meta-analysis were Chinese, with only one study on Canadian patients. Fourth, because of the different biological characteristics of the tumors and fact that most of the literature did not contain enough clinical data, the conclusion that TUSC7 is not correlated with tumor size, regional lymph node metastasis, distant organ metastasis or TNM staging requires further validation. Finally, although data in large scale suggest that TUSC7 is associated with the ubiquitination process, it remains to be proven through molecular biology experiments.
In conclusion, though with limitations, our results revealed that the expression of lncRNA TUSC7 is under-expressed in all sorts of cancer tissues, compared to normal tissues, and is significantly correlated with OS, and is potentially a prognostic marker of cancers. Genome-wide microarray and RNA-seq data suggest that TUSC7 has a potential regulatory role in ubiquitination. More comprehensive and large-scale clinical trials are required to elucidate the prognostic value of lncRNA TUSC7 expression in various cancers.
Full table
Acknowledgments
Funding: The study was supported by the Youth Scientific Project Foundation of Guangzhou Medical University (2015A21) and the Foundation for Distinguished Young Talents in Educational Commission of Guangdong Province of China (2016KQNCX128). This study was supported in part by the National Natural Science Foundation of China (81672270), Guangdong Province Natural Science Foundation (2015A030313474) and Key project of Guangzhou Science Technology and Innovation Commission (201707020042).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014;63:1700-10. [Crossref] [PubMed]
- Zhou M, Guo M, He D, et al. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med 2015;13:231. [Crossref] [PubMed]
- Shi D, Qu Q, Chang Q, et al. A five-long non-coding RNA signature to improve prognosis prediction of clear cell renal cell carcinoma. Oncotarget 2017; [Epub ahead of print].
- Zhou M, Zhong L, Xu W, et al. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci Rep 2016;6:31038. [Crossref] [PubMed]
- Yuan S, Wang J, Yang Y, et al. The Prediction of Clinical Outcome in Hepatocellular Carcinoma Based on a Six-Gene Metastasis Signature. Clin Cancer Res 2017;23:289-97. [Crossref] [PubMed]
- Wang Z, Jin Y, Ren H, et al. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Transl Res 2016;8:680-7. [PubMed]
- Wang Y, Liu Z, Yao B, et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol 2016;37:11429-41. [Crossref] [PubMed]
- Qi P, Xu MD, Shen XH, et al. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer 2015;137:1269-78. [Crossref] [PubMed]
- Tong YS, Zhou XL, Wang XW, et al. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med 2014;12:233. [Crossref] [PubMed]
- Qi P, Xu MD, Ni SJ, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med 2013;11:122. [Crossref] [PubMed]
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [Crossref] [PubMed]
- Rustici G, Kolesnikov N, Brandizi M, et al. ArrayExpress update--trends in database growth and links to data analysis tools. Nucleic Acids Res 2013;41:D987-90. [Crossref] [PubMed]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [Crossref] [PubMed]
- Pasic I, Shlien A, Durbin AD, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res 2010;70:160-71. [Crossref] [PubMed]
- Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res 2013;41:4976-87. [Crossref] [PubMed]
- Ding YC, Yu W, Ma C, et al. Expression of long non-coding RNA LOC285194 and its prognostic significance in human pancreatic ductal adenocarcinoma. Int J Clin Exp Pathol 2014;7:8065-70. [PubMed]
- Cong M, Li J, Jing R, et al. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol 2016;37:9441-50. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Lin A, Hu Q, Li C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol 2017;19:238-51. [Crossref] [PubMed]
- Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370-81. [Crossref] [PubMed]
- Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013;500:598-602. [Crossref] [PubMed]
- Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. N Engl J Med 2016;374:2287-9. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013;4:2939. [Crossref] [PubMed]
- Zhang A, Zhao JC, Kim J, et al. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep 2015;13:209-21. [Crossref] [PubMed]
- Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ 2017;24:59-71. [Crossref] [PubMed]