
Exosomal miRNAs in nipple aspirate fluid and breast cancer
Introduction
Exosomes are small vesicles with a lipid bilayer membrane containing a small cytosolic core. They can transport proteins, DNA, mRNA, miRNAs and lipids from one cell to another, thereby transferring genetic information and influencing the cells that they interact with (1). The content of exosomes may be different from the originating cell based on sorting of the contents within the exosome (1). Exosomes from malignant cells have been shown to induce neoplastic transformation of normal cells (2). Exosome mediated transfer of miR10b from MDA-MB-231 breast cancer cells increased the ability of immortalized human mammary epithelial cells to develop invasive properties (3). Exosomes isolated from the serum of breast cancer patients (but not those from healthy donors) induced tumor formation in mice when co-injected with nontumorigenic epithelial cells (4). Tumor-derived exosomes also mediate treatment resistance by transferring drug resistance proteins and miRNAs, by inducing efflux of or encapsulation of cytotoxic drugs, and/or by countering the effect of the drugs (1). Tumor cells appear to release a higher amount of exosomes than nonmalignant cells to influence their growth and spread (1).
Body fluid miRNAs, inflammation and cancer
Exosomes have been detected in multiple body fluids, including blood, milk, urine, saliva, pleural fluid, cerebrospinal fluid and the aqueous humor of the eye. Recently, exosomes from human saliva have been characterized for their mRNA content and 509 mRNAs were found (5). Exosomes containing miRs have been identified in serum from breast cancer patients (6). Exosomes have been identified in the urine of patients with bladder (7), lung (8) and prostate cancer (9). Exosomes isolated from the pleural effusions of lung cancer patients contain proteins related to lung cancer signaling (10). Other reports evaluating body fluids from noncancer patients document exosomes in the aqueous humor of the eye (11) and in cerebrospinal fluid (12).
We analyzed 10 miRNAs (16, 21, 100, 129, 145, 155, 181, 199, 205, 212) in nipple aspirate fluid (NAF) that have been associated with cancer. miR100 is reported to decrease the production of breast cancer stem cells, and its expression in tumors has been inversely correlated with patient survival (13). We previously observed higher expression of miR100 in normal tissue than in hormone sensitive rat mammary tumors (14). miR129 is thought to be a tumor suppressor which is frequently inactivated through methylation (15). Analysis of lymphoma and myeloma cell lines demonstrated methylation in all 13 samples analyzed (15). We observed differential expression (up in tumor but down in normal) in matched mammary tissue after treatment with resveratrol (14).
Downregulation of miR145 was reported to predict postmenopausal breast cancer risk (16). miR155 has been reported to increase cell plasticity and growth (17). When miR155 was overexpressed in B cells, it induced B cell malignancy (18). miR21 appears to play an important role in all phases of breast cancer pathogenesis (19). miR155 and miR21 have been implicated in breast cancer epithelial to mesenchymal transformation, cell migration and invasion control (17,20). Serum levels of miR16 (21), -21 (22), and -155 (23) have been reported to be significantly higher in patients with breast cancer than controls.
miR181 was evaluated in the serum of 88 patients with CRC and 11 healthy controls. Expression was significantly higher in patients with CRC (24). miR199 (14) and -204 (25) expression were found to be lower in mammary tumors than in matched controls.
miR205 is frequently detected in exosomes from body fluids. It can act both as a tumor suppressor and as an oncogene (26). As a tumor suppressor, miR205 acts as an inhibitor of cell proliferation, migration and invasion. On the other hand, as an oncogene, miR205 promotes tumor initiation and development. miR205 was measured in the serum of 58 breast cancer patients and 93 healthy controls. Levels in the serum of healthy women were higher (P<0.01) than in women with breast cancer (27). A second paper (28) observed the opposite trend, with higher levels of miR205 in the serum of women with breast cancer compared to controls. MCF7 derived exosomes have been reported to have low levels of miR205 (29).
miR212 has been shown to act a tumor suppressor, inhibiting the growth of lung cancer (30) and cervical cancer (31). Many of the same exosomal miRNAs that are present in cancer body fluids are also important in the regulation of inflammation (32), and chronic inflammation is a known risk factor for the development of cancer. We sought to determine if exosomes in NAF contain miRNAs, and the ability of the miRNA concentration to predict (I) breast cancer; and (II) response to treatment with the anti-inflammatory/anti-cancer agents vitamin (vit) D and celecoxib.
Methods
Participants and sample collection
Participants were enrolled in two Institutional Review Board approved studies. This project received ethics approval from the University of Missouri, IRB #1024344, and the University of North Dakota, IRB #200806-372. All participants gave informed consent before taking part. The first study involved 32 women ranging in age from 24–82 years who were undergoing diagnostic biopsy to determine if they had breast cancer. Twenty were recently diagnosed with breast cancer while 12 were found to have benign disease. Seven of the 20 participants with breast cancer had disease spread to their ipsilateral axillary lymph nodes. Four of the women had triple negative breast cancers, while most of the remainder were estrogen and/or progesterone receptor (ER/PR) positive (one ER/PR unknown). All participants had HER2 negative tumors (Table 1).
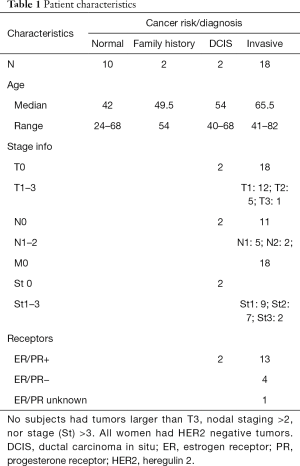
Full table
In the second study, matched NAF and serum samples were collected from five healthy women ranging in age from 33–61 years. For both studies, subjects were excluded if they had been diagnosed with cancer and had received definitive treatment (tumor removal, radiation, and or systemic therapy) prior to enrollment. After the participant provided informed consent, nipple fluid was aspirated by a trained physician or nurse clinician using a modified breast pump (33). NAF samples were collected in 50 micron capillary tubes, snap frozen and stored at −80 °C. The samples were batched until analysis. For women in the vitD/celecoxib study, blood was also collected, serum separated and snap frozen until use.
Isolation and resuspension of exosomes
An exosome isolation kit (System Biosciences Catalog # EXOQ5A-1, Mountain View, CA, USA) was used per manufacturer’s instructions. Briefly, NAF samples were diluted in 200 µL phosphate-buffered saline and exosome isolation reagent added, incubated at room temperature, centrifuged, and the excess supernatant removed. The samples were centrifuged a second time to remove additional supernatant. The exosome pellets were resuspended in 200 µL phosphate-buffered saline. The miRNA micro kit (Qiagen) was used to isolate RNA from exosomes per the manufacturer’s instruction. RNA was eluted in 14 µL H2O.
miRNA analysis in NAF and serum exosomes
We evaluated the expression in NAF of 10 miRNAs (16, 21, 100, 129, 145, 155, 181, 199, 205, 212). Matched serum was also analyzed in the vitD/celecoxib study. miRNA489, whose expression exhibited minimal variability between participants, was not significantly different in subjects with/without breast cancer nor significantly altered by vitD/celecoxib treatment, was used as a control. Primers were purchased from Qiagen (Valencia, CA, USA). Eight µL total RNA was used to generate cDNA using the miScript Reverse Transcription kit (Qiagen). The miScript SYBR Green PCR kit (Qiagen) was used in real time quantitative PCR for analysis of miRNA expression. RT and qPCR steps were followed according to the manufacturer’s instructions.
Statistical analysis
To evaluate the association of exosomal miRNAs in NAF with breast cancer, we compared expression in women with newly diagnosed breast cancer (invasive or ductal carcinoma in situ) with those without. Distributions for each of the miRNAs were first examined and found not to meet assumptions of normality needed for parametric analysis. Therefore, descriptive data are offered as median and interquartile range (IQR). Wilcoxon rank sum tests were conducted to compare the cancer and no cancer groups. Further analysis was done exploring node negative and positive groups within the cancer group. For the smaller sub-sample (less than 30) the exact test alternative was applied. There were no ties in the smaller data set.
Results
miRNA expression level in NAF exosomes does not significantly vary in NAF from cancer containing breasts vs. the breasts of women with benign disease, but changes with disease progression
Among women requiring diagnostic breast biopsy for a suspicious lesion found on breast imaging and/or physical exam, we did not observe a significant difference in the expression of 10 miRNAs analyzed in NAF (Table 2) comparing the cancer and noncancer groups. Among those with breast cancer, there was increased expression of miR16 and miR155 (Table 3) in women with node positive breast cancer compared to those with node negative disease (P=0.030 for both miRNAs).
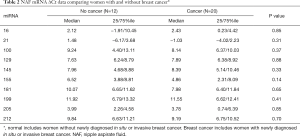
Full table

Full table
miRNA expression in NAF and serum exosomes in healthy women
Due to sample size limitations, we did not perform statistical analyses to comparing expression of the 10 miRNAs in NAF or serum before and after vitD +/− celecoxib treatment. There were no notable changes in expression after treatment (data not shown). On the other hand, miRNA expression was at different (defined at ∆Ct difference ≥1) in serum than matched NAF for all miRNAs, with higher levels in serum.
Discussion
One of the limitations of the current methods to diagnose breast cancer is the need for tumor tissue, which requires an invasive biopsy. Once obtained, a limitation of the tissue or cell sample is the intrinsic heterogeneity in breast cancer. Body fluid analysis minimizes the first limitation. NAF collection is totally noninvasive, whereas blood draw, for example, does involve needle access to a subcutaneous vein. Moreover, body fluid biomarkers are in concept a mix of each tumor cell’s biomarker expression pattern. As such, if one or more body fluid markers could be validated, they may be more robust than a tissue marker, since tissue sampling and analysis involves only a fraction of the entire heterogeneous tumor burden, increasing the chances that expression will vary from biopsy to biopsy.
We are not aware of a report documenting the detection of exosomes in NAF, nor the presence and measurement of miRNAs. We chose to analyze 10 miRNAs that have been associated with cancer, most with breast cancer (34,35). We were able to detect all 10 miRNAs in every NAF sample. Categorized based on whether the woman had newly diagnosed cancer or not, none were significantly associated with breast cancer.
Prior reports indicate that serum levels of miR16 (21) and -155 (23) are significantly higher in patients with breast cancer than controls. While we did not observe this trend in NAF exosomes, we did observe higher levels of these miRNAs in women with more advanced disease. Specifically, we observed significantly higher levels of miR16 and miR155 in women with node positive compared to those with node negative breast cancer. miRNAs are associated with normal functioning of the lymphatic system, alterations in miRNA155 decrease T and B cell normal functioning, and both aberrant miR16 and miR155 have been associated with lymphomagenesis (36). It is possible that higher levels of these miRNAs in the breast fluid encouraged tumor spread into the lymphatic system, resulting in disease progression.
In summary, we determined if exosomes were detectable in NAF, if the exosomes contained biologic information which could be reliably measured, and if the information obtained would be of potential clinical usefulness. We observed that exosomes are detectable, that they contain miRNAs associated with breast cancer, and that two of the miRNAs were associated with breast cancer progression through the lymphatic system.
Acknowledgments
We thank Ilene Staff, PhD, at Hartford Hospital for her assistance with statistical analysis.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Body Fluid Exosomes and Cancer”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.14). The series “Body Fluid Exosomes and Cancer” was commissioned by the editorial office without any funding or sponsorship. ERS served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Translational Cancer Research from Feb 2017 to Jan 2019. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the University of Missouri (No. #1024344) and the University of North Dakota (No. #200806-372). Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol 2015;8:83. [Crossref] [PubMed]
- Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014;32:983-97. [Crossref] [PubMed]
- Singh R, Pochampally R, Watabe K, et al. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer 2014;13:256. [Crossref] [PubMed]
- Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014;26:707-21. [Crossref] [PubMed]
- Record M, Subra C, Silvente-Poirot S, et al. Exosomes as intracellular signalosomes and pharmacological effectors. Biochem Pharmacol 2011;81:1171-82. [Crossref] [PubMed]
- Eichelser C, Stuckrath I, Muller V, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014;5:9650-63. [Crossref] [PubMed]
- Beckham CJ, Olsen J, Yin PN, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol 2014;192:583-92. [Crossref] [PubMed]
- Li Y, Zhang Y, Qiu F, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 2011;32:1976-83. [Crossref] [PubMed]
- Mitchell PJ, Welton J, Staffurth J, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med 2009;7:4. [Crossref] [PubMed]
- Park JO, Choi DY, Choi DS, et al. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics 2013;13:2125-34. [Crossref] [PubMed]
- Dismuke WM, Challa P, Navarro I, et al. Human aqueous humor exosomes. Exp Eye Res 2015;132:73-7. [Crossref] [PubMed]
- Tietje A, Maron KN, Wei Y, et al. Cerebrospinal fluid extracellular vesicles undergo age dependent declines and contain known and novel non-coding RNAs. PLoS One 2014;9:e113116 [Crossref] [PubMed]
- Deng L, Shang L, Bai S, et al. MicroRNA100 inhibits self-renewal of breast cancer stem-like cells and breast tumor development. Cancer Res 2014;74:6648-60. [Crossref] [PubMed]
- Qin W, Zhang K, Clarke K, et al. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutr Cancer 2014;66:270-7. [Crossref] [PubMed]
- Wong KY, Yim RL, Kwong YL, et al. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J Hematol Oncol 2013;6:16. [Crossref] [PubMed]
- Muti P, Sacconi A, Hossain A, et al. Downregulation of microRNAs 145-3p and 145-5p is a long-term predictor of postmenopausal breast cancer risk: The ORDET prospective study. Cancer Epidemiol Biomarkers Prev 2014;23:2471-81. [Crossref] [PubMed]
- Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015;5:1122-43. [Crossref] [PubMed]
- Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A 2006;103:7024-9. [Crossref] [PubMed]
- Zhang ZJ, Ma SL. miRNAs in breast cancer tumorigenesis Oncol Rep 2012;27:903-10. (Review). [Crossref] [PubMed]
- Han M, Liu M, Wang Y, et al. Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol Cell Biochem 2012;363:427-36. [Crossref] [PubMed]
- Hu Z, Dong J, Wang LE, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis 2012;33:828-34. [Crossref] [PubMed]
- Wu W, Sun C, Xu D, et al. Expression of CXCR2 and its clinical significance in human colorectal cancer. Int J Clin Exp Med 2015;8:5883-9. [PubMed]
- Roth C, Rack B, Muller V, et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res 2010;12:R90. [Crossref] [PubMed]
- Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921 [Crossref] [PubMed]
- Wang X, Qiu W, Zhang G, et al. MicroRNA-204 targets JAK2 in breast cancer and induces cell apoptosis through the STAT3/BCl-2/survivin pathway. Int J Clin Exp Pathol 2015;8:5017-25. [PubMed]
- Vosgha H, Salajegheh A, Smith RA, et al. The important roles of miR-205 in normal physiology, cancers and as a potential therapeutic target. Curr Cancer Drug Targets 2014;14:621-37. [Crossref] [PubMed]
- Zhang H, Li B, Zhao H, et al. The expression and clinical significance of serum miR-205 for breast cancer and its role in detection of human cancers. Int J Clin Exp Med 2015;8:3034-43. [PubMed]
- Shaker O, Maher M, Nassar Y, et al. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene 2015;560:77-82. [Crossref] [PubMed]
- Guzman N, Agarwal K, Asthagiri D, et al. Breast Cancer-Specific miR Signature Unique to Extracellular Vesicles Includes "microRNA-like" tRNA Fragments. Mol Cancer Res 2015;13:891-901. [Crossref] [PubMed]
- Luo J, Meng C, Tang Y, et al. miR-132/212 cluster inhibits the growth of lung cancer xenografts in nude mice. Int J Clin Exp Med 2014;7:4115-22. [PubMed]
- Zhao JL, Zhang L, Guo X, et al. miR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life 2015;67:380-94. [Crossref] [PubMed]
- Alexander M, Hu R, Runtsch MC, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun 2015;6:7321. [Crossref] [PubMed]
- Sauter ER, Daly M, Linahan K, et al. Prostate-specific antigen levels in nipple aspirate fluid correlate with breast cancer risk. Cancer Epidemiol Biomarkers Prev 1996;5:967-70. [PubMed]
- Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012;31:1609-22. [Crossref] [PubMed]
- Suzuki H, Maruyama R, Yamamoto E, et al. DNA methylation and microRNA dysregulation in cancer. Mol Oncol 2012;6:567-78. [Crossref] [PubMed]
- Lawrie CH. MicroRNAs and lymphomagenesis: a functional review. Br J Haematol 2013;160:571-81. [Crossref] [PubMed]


