
Expression of HGF, MET and mutated EGFR in EGFR mutated lung adenocarcinoma and their clinical significance
Introduction
Lung cancer is the most common cancer worldwide, and is also the main cause of cancer-related deaths (1), wherein non-small cell lung cancer (NSCLC) accounts for about 80%, and the 5-year survival rate is only about 15% (2). Currently, approximately 70% of patients with NSCLC are already in the advanced stage when diagnosed and lose the opportunity for surgery, therefore, radiotherapy and chemotherapy are the main treatments for them. Patients with advanced NSCLC who have epidermal growth factor receptor (EGFR)-sensitive mutations are highly sensitive to EGFR-tyrosine-kinase inhibitors (TKIs) with an overall response rate (ORR) of up to 70%, the median progression-free survival (PFS) is 9–13 months, and the overall survival (OS) is 20–30 months (3-6). Nevertheless, treatment-effective patients have different individual responses to drugs, and inevitably develop secondary drug resistance. The mechanisms of acquired drug resistance include EGFR T790M mutation, mesenchymal epithelial transition (MET) or HER2 amplification, epithelial-mesenchymal transition (EMT), transformation of small cell lung cancer, etc. (7-10). Furthermore, about 20–30% patients with EGFR-sensitive mutations present primarily resistant to EGFR-TKIs, and the possible causes include tumor heterogeneity, abundance of EGFR mutations, genetic polymorphism of Bcl-2 interacting mediator of cell death (BIM), bypass activation such as MET and PI3K/AKT pathway, and co-mutations with de-novo EGFR T790M or KRAS.
Recently, studies have shown that abnormal activation of hepatocyte growth factor (HGF)/MET signaling pathways may play an important role in both primary and secondary drug resistance to EGFR-TKIs (10-13), and these patients tend to have a poor and short duration of response to EGFR-TKIs (12,14). Therefore, in the present study, we analyzed the correlation between clinicopathological characteristics of lung cancer and heterogeneous expression of HGF and MET proteins, the type of EGFR-sensitive mutations and their protein expression, as well as their relationship with recurrence and survival, and further investigated whether additionally detecting the expression of HGF and MET proteins could accurately predict the prognostic survival and efficacy for patients with EGFR-sensitive mutations treated with EGFR-TKIs.
Methods
Clinical data
A total of 130 patients with lung adenocarcinoma were screened for EGFR mutations of exon 19 (E19) deletion (del) and exon 21 (E21) L858R by the amplification refractory mutation system (ARMS) method from May 2005 to April 2014 in Shengjing Hospital of China Medical University. All patients had complete clinical and pathological data available. Pathological tissue was obtained from surgical resection in 80 cases, and from computed tomography (CT) or ultrasound-guided or bronchoscopic aspiration biopsy in 50 cases. All patients had not received chemotherapy and radiotherapy prior to pathological diagnosis. Postoperative patients were given adjuvant chemotherapy and radiotherapy according to their clinicopathological staging. The cutoff date for patient follow-up was March 20, 2015. Histopathological classification was in accordance with the World Health Organization standards, while Tumor Node Metastasis staging was with the AJCC Cancer Staging Manual, 7th Edition.
Immunohistochemistry of E19 del/E21 L858R-EGFR, HGF and MET expression
Rabbit anti-human E19 del (#2085) and E21 L858R (#3197) protein antibodies were purchased from Cell Signaling Technology (San Francisco, CA, USA), rabbit anti-human MET (sc-10) and HGF (sc-7949) protein antibodies were purchased from Santa Cruz Biotechnology (Delaware Ave, Santa Cruz, CA, USA), and universal secondary antibodies (ZSGB-BIO, SP-9001, Beijing, China) were gifted by the Pathology Laboratory of Shengjing Hospital. E19 del or E21 L858R diseased tissue specimens were fixed by 10% formalin solution and paraffin-embedded as 4 µm serial sections. The pathological sections of the patients with mutations were respectively subjected to S-P immunohistochemistry staining by using HGF, MET antibodies and the corresponding E19 del or E21 L858R protein antibodies. Specific steps were as follows. The paraffin sections were dewaxed with gradient alcohol and xylene then water, placed in 3% hydrogen peroxide and incubated at room temperature for 10 min to remove endogenous peroxidase activity. A microwave was used to hot fix antigen for 3 min, 3.5–10% goat serum was used to seal non-specific sites, then incubated at room temperature for 10 min. Serum was decanted and samples incubated overnight at 4 °C in diluted primary-antibody solution (MET and E21 antibodies at 1:75 dilution, and HGF and E19 antibodies at 1:100 dilution). Working solutions of biotin-labeled secondary-antibody and horseradish peroxidase-labeled streptavidin-peroxidase were added consecutively, and each step was incubated in a wet box for 20 min. Diaminobenzidine staining, hematoxylin re-staining and mounting were then carried out.
Assessment of immunohistochemistry results
Cytoplasm or cell membrane as clearly yellowish-brown granules was suggestive of positive expression, where E19 del or E21 L858R EGFR protein and MET protein were mainly expressed in tumor cells, while HGF protein was expressed in both tumor cells and stromal cells. After minor modification of a previous method (12), five fields were observed at random under high power lens (×400), and semi-quantitative analysis was conducted based on the percentage of positive cells and staining intensity. Staining intensity was scored as: 0 point for no staining, 1 point for mild staining, 2 points for moderate staining, and 3 points for severe staining; 0 point for positive cells <10%, 1 point for 10–25%, 2 points for 25–50%, and 3 points for >50%. If the sum of the two indicators was 0–1 point, the case was negative (−); if the sum was 2–3 points, the case was weakly positive (+); if the sum was ≥4 points, the case was strongly positive (2+). Weakly positive and strongly positive were collectively referred to as positive.
Statistical analysis
IBM SPSS statistic version 19.0 (IBM Corporation, New York, USA) was employed for statistical analysis. Chi-square test and Fisher exact test were used for inter-group and intra-group analysis, survival curve was drawn using the Kaplan-Meier method, and log-rank test was adopted to compare survival differences. Cox’s proportional hazards regression model was employed for multivariate analysis to determine the prognostic factors. P<0.05 indicated statistically significant differences.
Results
Protein expression of EGFR E19 del or E21 L858R protein, and HGF and MET
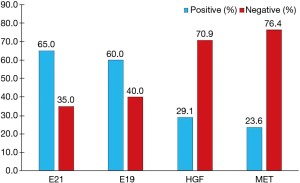
According to the integrity status of pathological tissue and the following detection of EGFR mutation using the ARMS method, eventually the samples of 55 patients (60%) with EGFR E19 or E21-sensitive mutations underwent immunohistochemistry. The clinicopathological characteristics were summarized in Table 1, including gender, age, smoking status, stage, differentiation and EGFR mutation type. Among them, 50 cases were pathologically confirmed after surgical resection and five cases by biopsy. There were 35 patients with E19 del mutations and 20 patients with E21 L858R mutations, respectively, in 19 males and 36 females with a median age of 59 years. Immunohistochemistry results showed cytoplasm or cell membrane staining with yellowish-brown granules was suggestive of positive expression. EGFR E19 del or E21 L858R protein and MET protein were mainly expressed in tumor cells, while HGF protein was expressed in both tumor cells and stromal cells. Immunohistochemistry results of EGFR E19 del and E21 L858R protein expression are shown in Figure 1, and the results of HGF and MET proteins are shown in Figure 2. Overall, 21/35 cases (60.0%) with E19 del and 13/20 cases (65.0%) with E21 L858R were positive for corresponding specific mutant protein expression; 16/55 cases (29.1%) were positive for HGF protein expression; and 13/55 cases (23.6%) were positive for MET protein expression, as shown in Figure 3.
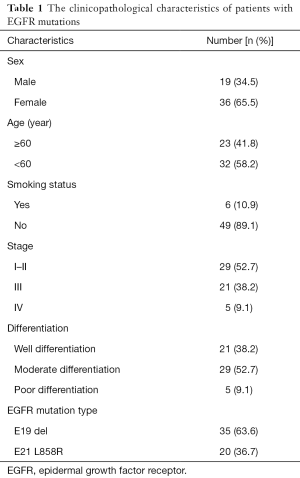
Full table
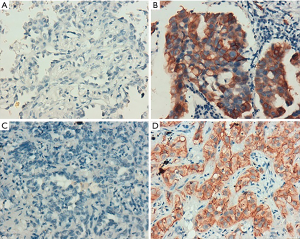
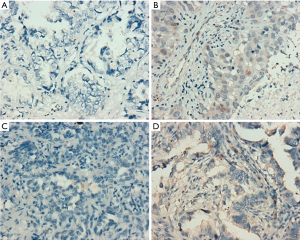
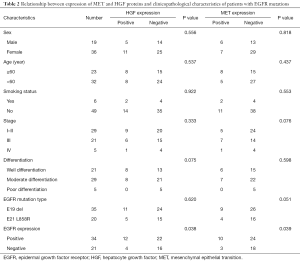
Relationship between EGFR mutations and protein expression of MET and HGF, and clinicopathological characteristics of patients
In the 55 cases with EGFR mutations, HGF and MET protein expression were positively correlated with EGFR-mutant protein expression (P=0.038, r =0.068; P=0.039, r =0.065, respectively), but both were irrelevant to the type of EGFR mutations (P>0.05). The expression of EGFR-mutant proteins was not associated with clinicopathological characteristics (all P>0.05). Meanwhile, the expression of HGF and MET proteins was not correlated with other clinicopathological characteristics including sex, age, smoking status, stage and degree of differentiation of patients (all P>0.05), as shown in Table 2.
Full table
Relationship between expression of HGF proteins and survival
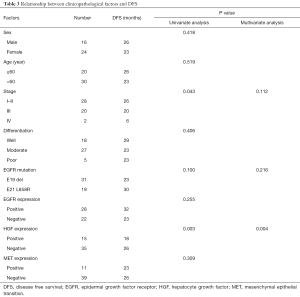
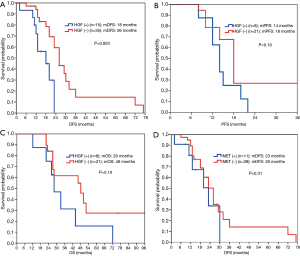
The 55 patients with EGFR sensitive mutations were followed up for a median period of 48 months. Twenty-eight of the patients (50.9%) experienced lung cancer-related death. Median OS was 42 months. Among them, 50 patients who underwent radical resection did not receive postoperative EGFR-TKI treatment, and 29 patients who either experienced recurrence after surgery or were initially diagnosed with advanced disease, received EGFR-TKI treatment. Univariate analysis showed that HGF expression and stage were associated with disease-free survival (DFS) in postoperative patients, while multivariate analysis showed that only HGF expression was an independent prognostic factor of DFS for postoperative patients (P=0.004, Table 3). Patients with negative HGF expression had significantly improved DFS (26 vs. 18 months, P=0.003) compared to patients with positive HGF expression (Figure 4A). For 29 patients with recurrent or advanced NSCLC who received first or later lines of EGFR-TKI treatment, ORR in the HGF expression-negative group (76%) was significantly higher than that in the positive group (25%) (P=0.035), as shown in Table 4. Both PFS (18 vs. 14 months, P=0.19) and OS (48 vs. 28 months, P=0.10) were prolonged in HGF expression-negative group compared to the HGF expression-positive group, but without significant difference (Figure 4B,4C).
Full table

Full table
Relationship between expression of MET proteins and survival
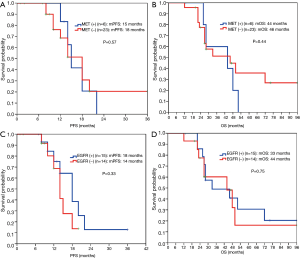
The expression level of MET protein was irrelevant to DFS for postoperative patients (Figure 4D). No significant differences were found in PFS (18 vs. 15 months, P=0.57) and OS (46 vs. 44 months, P=0.44) between MET expression-negative group and positive group for patients who received EGFR-TKI treatment (Figure 5A,5B).
Relationship between expression of EGFR-mutant proteins and survival
No significant differences were found in PFS (14 vs. 18 months, P=0.33) and OS (44 vs. 33 months, P=0.75) between EGFR-mutant protein expression-negative group and positive group for patients who received EGFR-TKI treatment (Figure 5C,5D).
Discussion
Studies have shown that HGF can mediate the occurrence of secondary drug resistance to EGFR-TKIs in patients with EGFR mutant lung cancer (11-13,15). A Japanese study (16) indicated that the HGF protein was expressed in some patients with primary drug resistance to EGFR-TKIs, suggesting that HGF may have been involved in primary drug resistance apart from secondary drug resistance to TKIs, though this is currently not very clear. The present study showed that HGF protein expression was an independent prognostic factor affecting DFS in postoperative patients with lung adenocarcinoma (P=0.004, Table 3), and patients with HGF expression-negative had a significantly higher DFS than patients with HGF expression-positive (26 vs. 18 months, P=0.003, Figure 4A). Patients with HGF expression-negative had a significantly better ORR (76% vs. 25%, P=0.035, Table 4) and tended to survive longer than patients with HGF expression-positive (PFS: 18 vs.14 months, P=0.19; OS: 48 vs. 28 months, P=0.10, Figure 4B,4C). Therefore, for patients with advanced lung adenocarcinoma who harbored EGFR-sensitive mutations and received EGFR-TKI treatment, additional detection of HGF protein expression may be useful for further accurately predicting efficacy and survival.
HGF, as a natural ligand of MET, activates downstream signaling pathways by binding to MET, and promotes cell proliferation. Recent studies have shown that HGF may cause drug resistance to EGFR-TKIs in patients with EGFR-mutant lung cancer by activating a bypass of MET/Gab1/PI3K/AKT or accelerating clonal growth of tumor cells carrying the MET gene amplification (11,17). In this study, the positive rate of HGF protein expression in EGFR-TKIs untreated patients with EGFR mutations was only 29.1% (Figure 3), consistent with the positive rate in patients with primary drug resistance reported in the literature, but lower than the positive rate of HGF protein expression of 61% after drug resistance reported in the literature (16). One of the reasons may be that HGF in tumor stroma in untreated patients remains in a low-expression status, but after EGFR-TKI treatment, the tumor microenvironment changes, thus secreting large amounts of HGF. However, in the present study, re-biopsy was not conducted after drug resistance to EGFR-TKIs, so changes in HGF expression before and after drug resistance could not be compared. Another reason may be related to the earlier staging of patients in this study, but there was no significant difference in the expression of HGF protein among patients with different stages.
More importantly, we found that patients who received EGFR-TKI treatment had a significantly higher ORR (76% vs. 25%, P=0.035, Table 4) in the HGF expression-negative group than the expression-positive group. OS in HGF expression-positive patients tended to be worse than in HGF expression-negative patients (28 vs. 48 months, P=0.10, Figure 4C). Multivariate analysis showed that stage and HGF expression were independent prognostic factors of OS in patients with advanced disease receiving EGFR-TKI treatment (P=0.023 and 0.048). Our results suggested that HGF expression might be an important biomarker used to predict poor response and survival after EGFR-TKI treatment. Therefore, additional detection of HGF protein expression in patients with EGFR-sensitive mutations could further accurately predict the efficacy of EGFR-TKIs and prognostic survival. Furthermore, for postoperative patients, an improved DFS was observed in patients with negative HGF expression compared to patients with positive HGF expression (26 vs. 18 months, P=0.003, Figure 4A). Multivariate analysis also showed that HGF protein expression was an independent prognostic factor for survival (P=0.004, Table 3). Nevertheless, different subsequent treatments after failure of EGFR-TKI treatment were given to some patients, including chemotherapy, radiotherapy, symptomatic and supportive treatment. Because of the limited sample size, we did not further analyze the possible influence of these treatments on survival. Future studies with larger numbers of patients are needed to confirm these findings.
MET-induced drug resistance is related to MET activation by a two way-receptor-ligand activation pathway and a dimeric MET activation pathway caused by HGT later binding to MET (18,19), activating a variety of intracellular signaling pathways, such as PI3K/AKT, MAPK and Stat3, thus causing the proliferation, invasion and metastasis of tumors. A previous study has shown that MET gene amplification is positively correlated with MET protein expression (20). This study found positive MET protein expression in 13/55 patients (23.6%), as shown in Figure 3. A meta-analysis suggested that the prognosis in patients with higher MET protein expression was relatively poor (21). This study showed no significant difference in the impacts of heterogeneous MET protein expression on the survival and prognosis of patients. The results were inconsistent with previous literature, which may be related to the small number of cases in this study. Meanwhile HGF and MET expressions were not completely consistent in terms of space and amount of expression, maybe because MET-induced drug resistance caused the different expressions of these two proteins through the above mentioned two way-receptor-ligand activation pathway and dimeric MET activation pathway caused by HGT after binding to MET.
Another study has shown (22) that the response to EGFR-TKIs was better and PFS was longer in patients with higher mutant protein expression than in patients with lower mutant protein expression. One study (23) showed that EGFR-mutant protein expression may be correlated to mutation abundance to a certain extent, that is, the higher the mutation abundance, the higher the protein expression levels, which can thus better predict the efficacy. This study found that the positive rate of EGFR-mutant protein expression was only 65% in patients with known E19 or E21 mutations, consistent with literature reports (24,25). The present study found that in patients with advanced lung cancer receiving EGFR-TKIs, there was no statistical difference in PFS between EGFR-mutant protein expression-positive patients and expression-negative patients (18 vs. 14 months, P=0.33, Figure 5C). The reason may be that we found the positive rate of HGF and MET protein expression in patients with positive mutant EGFR protein expression was significantly higher than in patients with negative expression. Thus, at least in the EGFR-mutation-positive lung cancer population, EGFR mutated protein may not further predict the efficacy or disease progression after EGFR-TKI treatment.
In conclusion, the HGF protein is an independent prognostic factor affecting the early recurrence for postoperative patients with EGFR-mutant lung adenocarcinoma. For advanced patients receiving EGFR-TKIs, HGF may be involved in primary drug resistance, and is also an independent predictor of the efficacy of EGFR-TKIs and survival. Therefore, additional HGF protein detection may better predict the efficacy of EGFR-TKIs and prognostic survival in patients with EGFR mutations. However, due to the limited sample size in our study, there may be bias, so future studies with larger sample sizes are needed to validate these findings.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (grant No. 81501990) and the Talent Support Project of Colleges and Universities of Liaoning Province (grant No. LJQ2014082).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.44). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Wistuba II, Gelovani JG, Jacoby JJ, et al. Methodological and practical challenges for personalized cancer therapies. Nat Rev Clin Oncol 2011;8:135-41. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol 2015;26:1877-83. [Crossref] [PubMed]
- Inoue A, Kobayashi K, Maemondo M, et al. Updated overall survival results from a randomized phase III trial comparing gefitinib with carboplatin-paclitaxel for chemo-naive non-small cell lung cancer with sensitive EGFR gene mutations (NEJ002). Ann Oncol 2013;24:54-9. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73 [Crossref] [PubMed]
- Popat S, Wotherspoon A, Nutting CM, et al. Transformation to “high grade” neuroendocrine carcinoma as an acquired drug resistance mechanism in EGFR-mutant lung adenocarcinoma. Lung Cancer 2013;80:1-4. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res 2008;68:9479-87. [Crossref] [PubMed]
- Yamada T, Takeuchi S, Kita K, et al. Hepatocyte growth factor induces resistance to anti-epidermal growth factor receptor antibody in lung cancer. J Thorac Oncol 2012;7:272-80. [Crossref] [PubMed]
- Matsumoto K, Nakamura T. Hepatocyte growth factor and the Met system as a mediator of tumor-stromal interactions. Int J Cancer 2006;119:477-83. [Crossref] [PubMed]
- Raghav KP, Gonzalez-Angulo AM, Blumenschein GR Jr. Role of HGF/MET axis in resistance of lung cancer to contemporary management. Transl Lung Cancer Res 2012;1:179-93. [PubMed]
- Yamada T, Matsumoto K, Wang W, et al. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin Cancer Res 2010;16:174-83. [Crossref] [PubMed]
- Yano S, Yamada T, Takeuchi S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol 2011;6:2011-7. [Crossref] [PubMed]
- Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. [Crossref] [PubMed]
- Goyal L, Muzumdar MD, Zhu AX. Targeting the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res 2013;19:2310-8. [Crossref] [PubMed]
- Trovato M, Torre ML, Ragonese M, et al. HGF/c-met system targeting PI3K/AKT and STAT3/phosphorylated-STAT3 pathways in pituitary adenomas: an immunohistochemical characterization in view of targeted therapies. Endocrine 2013;44:735-43. [Crossref] [PubMed]
- Dziadziuszko R, Wynes MW, Singh S, et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol 2012;7:340-7. [Crossref] [PubMed]
- Guo B, Cen H, Tan X, et al. Prognostic value of MET gene copy number and protein expression in patients with surgically resected non-small cell lung cancer: a meta-analysis of published literatures. PLoS One 2014;9:e99399 [Crossref] [PubMed]
- Azuma K, Okamoto I, Kawahara A, et al. Association of the expression of mutant epidermal growth factor receptor protein as determined with mutation-specific antibodies in non-small cell lung cancer with progression-free survival after gefitinib treatment. J Thorac Oncol 2012;7:122-7. [Crossref] [PubMed]
- Okabe T, Okamoto I, Tamura K, et al. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res 2007;67:2046-53. [Crossref] [PubMed]
- Li AR, Chitale D, Riely GJ, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn 2008;10:242-8. [Crossref] [PubMed]
- Macarenco RS, Uphoff TS, Gilmer HF, et al. Salivary gland-type lung carcinomas: an EGFR immunohistochemical, molecular genetic, and mutational analysis study. Mod Pathol 2008;21:1168-75. [Crossref] [PubMed]


