
Circular RNAs in cancer: old tree with new flowers
Introduction
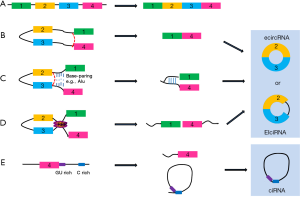
Circular RNAs (circRNAs) are a large group of RNAs predominantly generated by a process known as back splicing. Different from canonical linear isoforms, a splice donor site is joined to a splice accepter site further upstream in the primary transcript, yielding a covalently closed loop structures (Figure 1) (1). CircRNAs have long been supposed as a type of by-products of mis-splicing events due to the technical limitations or low levels of expression (2). Novel bioinformatic approaches coupled with biochemical enrichment strategies and deep sequencing have allowed comprehensive studies of circRNA species. Recent studies have revealed thousands of endogenous circRNAs in mammalian cells, most of which are stable and conserved (3,4).
Like microRNA (miRNA), circRNAs act as co- and post-transcriptional regulators for the modulation of gene expression in several pathogenic states (5), including carcinogenesis. Genes located in the center of several cancer-associated pathways are regulated by the circRNA-miRNA-mRNA or circRNA-mRNA axis (6). Therefore, clarification roles of circRNAs in regulating cancer biological development would have great potential significance (7,8). In particular, circRNAs have been shown more stable than their liner counterparts in plasma and saliva, indicating their potential as new clinical diagnostic and prognostic biomarkers. In this review, we will focus on novel insights into the functions of circRNAs, and discuss their potential significance and implications in cancer.
CircRNA function
CircRNA is a large class of RNAs with as-yet-totally undetermined functions and great diversities in length, from hundreds to thousands of nucleotides. How circRNAs play the pivotal regulatory role in gene expression is associated with their compositions and locations in cells.
CircRNAs composed of exons (ecRNAs) primarily reside in the cytoplasm and possibly have miRNA response elements (MREs) (4,9). Due to MREs, circRNAs ‘sponge up’ miRNAs and suppress the ability of miRNAs to bind their mRNA targets (10), thus reducing the effect of miRNA-mediated posttranscriptional repression and playing a regulatory role in competing endogenous RNA activities (11,12). The most notable circRNA is CDR1as/ciRS-7 (encoded in the genome antisense to the human CDR1 gene locus), harboring 74 miR-7 seed matches. It has been experimentally validated that over-expressing CDR1as/ciRS-7 increases the expression of miRNA target reporter constructs, while knocking down these circRNAs produces the opposite effect. The overexpression of CDR1as in zebrafish embryos, which lack the cdr1 locus, substantially reduces the midbrain size and mimics the phenotype of miR-7 loss-of-function, which causes morphological defects in the midbrain (4,12). Although these results indicate that some circRNAs indeed function as miRNA sponges, whether it is a common phenomenon and how networks of circRNAs and miRNAs maintain balance to regulate cellular homeostasis remain to be clarified.
Some circRNAs from introns (ciRNAs), or exons with introns ‘retained’ between exons (EIciRNAs) are primarily located in the nucleus in eukaryotes and regulate host gene expression (13,14). They share the common feature of little enrichment in miRNA target sites and predominant abundance in the nucleus, where they cis-regulate their parent genes through specific RNA-RNA interactions at gene promoter (13). Intron-derived circRNAs (ciRNA) demonstrated their capacity to enhance transcription of their host genes by interacting with Pol II machinery (14). EIciRNAs (exon-intron circRNAs), such as circEIF3J and circPAIP2, with complementary sequence pairing between EIciRNA and U1 snRNA, have analogous effects on parent gene transcription by interacting with U1 snRNP and Pol II (13). Further experiments revealed that knocking them down could significantly decrease their parent genes (5).
Besides regulating miRNAs, circRNA may bind and sequester RNA-binding proteins (RBPs), resulting in the formation of large RNA-protein complex (RPCs), probably by acting as competing elements in the same way as they modulate miRNAs. The unique tertiary structure of Some ecRNAs facilitate its storage, classification and localization of RBPs, such as Argonaute, RNA polymerase and MBL (15).
In addition to the above, a special function of circRNAs is the mRNA trap, which refers to the phenomenon of circRNA formation competing with productive linear mRNAs (16). One version of explanation is the competition between forward splicing and back splice. And a negative correlation is also found between formation of some circRNAs and their mature linear RNAs. Another speculation is that circRNAs act as the ‘mRNA trap’ by sequestering the translation start site, leaving a non-coding linear transcript and thereby reducing the expression level of the formin protein (16). For example, in human fibroblasts, 34% single exon circles contain a translation start compared with only 14% of all exons containing a translation start (17).
Landscape of circRNAs in cancer
Some primitive studies have evidenced that circRNA involved in carcinogenesis. For instance, by means of highly sensitive RNA sequence of normal and cancerous tissues identification, a large number of circRNA were identified differentially expressed in basal cell carcinoma (GEO accession: GSE74756) (18), glioblastoma (GEO accession: GSE86202), liver cancer (GEO accession: GSE78520), gastric cancer (GEO accession: GSE78092), pancreatic ductal adenocarcinoma (GEO accession: GSE69362/GSE79634) (19), and cutaneous squamous cell carcinoma (GEO accession: GSE74758) (20). Lately, a new kind of circRNA, fusion-circRNAs (f-circRNA), aberrant generated from cancer-associated chromosomal translocations that juxtapose distant complementary repetitive intronic sequences, may also contribute to tumor progression (21). All of these evidences push us to further understand the underlying role of circRNAs in malignant biological behavior of cancer cells.
Growing evidence in the past 10 years has suggested that abnormal expression of miRNAs is a sign of malignant tumor occurrence (22,23). By competing for binding to miRNAs, circRNAs indirectly play a vital role in the initiation, development and prognosis of cancer. Ample evidence has shown that circRNAs play critical roles in fine-tuning the level of miRNA-mediated regulation of gene expression by sequestering miRNAs involved in carcinogenesis (8,24). As we all know, there are three kinds of miRNAs in carcinogenesis: oncomiR, tumor suppressive miRNA, and context-dependent miRNA (pleiotropic miRNA) (25). Based on this understanding and knowledge about the crosstalk between circRNAs and miRNAs, we speculate that circRNAs may also play different roles in tumorigenesis and progression (Table 1).

Full table
Tumor suppressive circRNAs in cancer
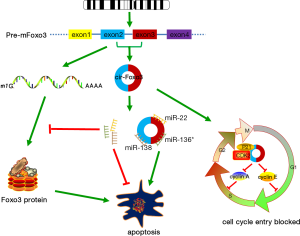
In certain situation, the reduction or deletion of a circRNA that functions as a tumor suppressor would lead to tumorigenesis. The overall outcome might involve the increased proliferation, invasiveness or angiogenesis, and decreased levels of apoptosis, or de-differentiation, ultimately leading to tumor formation. Anna Bachmayr-Heyda found a general principle that the ratio of circular to linear RNA isoforms was always lower in colorectal cancer (CRC) cell lines and cancer tissues compared with normal tissues. In addition, they revealed a negative correlation between global circular RNA abundance and the proliferation of cancer cells. Hsa_circ_0001649 was significantly down-regulated in HCC, and knocking down hsa_circ_0001649 with siRNAs in immortalized hepatoma cell lines significantly increased the mRNA levels of MMP9, MMP10 and MMP13, indicating that a low level of hsa_circ_0001649 expression was positively correlated with the metastasis of HCC (26). Remarkably, Li et al. found that cir-ITCH expression was usually low in esophageal squamous cell carcinoma (ESCC) compared with the peritumoral tissue. Cir-ITCH may play an antitumor role in ESCC by interacting with miRNAs such as miR-7, miR-17, and miR-214 and increased the concentration of ITCH, thus promoting ubiquitination and degradation of phosphorylated Dvl2 by inhibiting the Wnt/β-catenin pathway (27). Circ-Foxo3 shared 25 identical sequences with Foxo3 to bind miR-22, miR-136*, miR-138, miR-149*, miR-433, miR-762, miR-3614-5p and miR-3622b-5p, all of which could decrease cellular apoptosis. Ectopic expression of cir-Foxo3 in breast carcinoma cell MDA-MB-231 suppressed tumor growth, cancer cell proliferation and survival, not only as a consequence of their sponging effect on these miRNAs but the freedom of Foxo3 mRNA bound by these miRNAs to perform its own tumor suppressor function (28). Further study revealed that ectopic expression of circ-Foxo3 repressed cell cycle progression by binding to the cell cycle proteins cyclin-dependent kinase 2 (CDK2), and cyclin-dependent kinase inhibitor 1 (or p21), resulting in the formation of a ternary complex, which arrested the function of CDK2 and blocked cell cycle G1-S transition (Figure 2) (29).
Oncogenic circRNAs in cancer
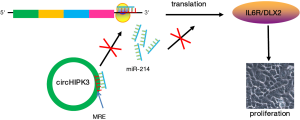
On the contrary, the amplification or overexpression of some circRNAs that have oncogenic roles would also lead to tumor formation. In this circumstance, increased amounts of certain circRNA, which might be produced at inappropriate times or in wrong tissues, would eliminate the suppression of a miRNA-target tumor-promotion gene and lead to cancer progression. Despite the above overwhelming evidence that supports the tumor suppressive role of circRNAs, the opposite effect has also been reported. For example, cZNF292, found to be expressed during hypoxic condition, promoted tube formation by increasing glioma cell proliferation and cell cycle progression. cZNF292 silencing blocked Cell cycle progression in human glioma U87MG and U251 cells halted at S/G2/M phase via the Wnt/β-catenin signaling pathway and related genes such as PRR11, Cyclin A, p-CDK2, VEGFR-1/2, p-VEGFR-1/2 and EGFR (30). In bladder cancer, circTCF25 significantly higher expressed in carcinoma tissue, could harbor and down-regulate miR-103a-3p and miR-107, increase CDK6 expression, and promote proliferation and migration in vitro and in vivo (31). CircHIPK3, derived from HIPK3 exon2, preferentially localized in the cytoplasm, was significantly upregulated in liver cancer compared with the matched normal tissue. Ectopic expression of circHIPK3 could attenuate the anti-proliferative effect of miR-124 by the up-regulated miR-124 proliferation-promoted targets IL6R and DLX2 (Figure 3) (32). In CRC tissues, circ_001569 was up-regulated and correlated with aggressive characteristics of CRC. Expression level was increased along with distant metastasis and poor differentiation. Gain-of-function and loss-of-function assays in SW620 and LOVO cells revealed that hsa_circ_001569 acted as a positive regulator in cell proliferation and invasion of CRC with sequestering miR-145 to terminate regulation of its target genes E2F5, BAG4 and FMNL2 (33).
Context dependent circRNAs in cancer
Last but not the least, some miRNAs, such as miR-7, have been characterized as having both oncogenic and tumor-suppressive properties (7,34). The relative circRNAs may also have pleiotropic roles. Taking ciRS-7 for example, on one hand, ciRS-7 strongly suppressed the miR-7 activity, increasing the level of miR-7 targets including oncogenic factors in cancer-related signaling pathways, such as EGFR (35), suggesting a distinct tumor promoting role of ciRS-7. On the other hand, miR-7 seems to be an oncogenic miRNA in HELA and lung carcinoma cell line, where ciRS-7 sponge up miR-7 may restrain tumor formation. In addition, Davide Barbagallo demonstrated that, ciRS-7 was a direct target of miR-671-5p, which over expression significantly increases migration and to a less extent proliferation rates of GBM cells. The axis miR- 671-5p/ciRS-7/CDR1/VSNL1 is functionally altered in GBM cells and is involved in the modification of their biopathological profile, suggesting tumor-suppressive properties of ciRS-7. Although there are few report firmly established the tumor suppressive function of ciRS-7, we still believe that it could be found out someday, but whether and how it works remain to be further investigated.
Particularly, our current knowledge about the roles of circRNAs in tumors may only catch a glimpse of the iceberg. The apparently contradictory functions as mentioned above may well be explained by an ambiguous role of their target genes, such as miRNAs, in regulating the complex networks.
CircRNAs as novel biomarkers
CircRNAs have been described as biomarkers of aging in Drosophila (36) and disease biomarkers in human saliva (37). Knowing that a large number of circRNAs are associated with cancer, it is possible to use certain types of circRNAs to improve the diagnosis of cancer. Additionally, there are several advantages of making them promising biomarkers for cancer. First, Most circRNAs enjoy extraordinary stability, likely as a result of covalently closed loop structures with neither 5’-3’ polarity nor a polyadenylated tail that are resistant to debranching enzymes (DBE) and RNA exonucleases (REN) as compared with their liner ones (3). Second, they can be secreted from the tissue to some types of extracellular body fluid. Small “free” circRNAs could be sorted into exosomes and exported out of cells. Exosomes enriched (at least by 2 folds) with stable circRNAs are moderately associated with cellular circRNAs. Compared with the content of linear RNAs detected in the human blood, circRNAs were found to be more stable in serum samples for at least 24 h at room temperature (38). Researchers have identified more than 400 circRNAs in human cell-free saliva from healthy individuals (39). Further studies have identified more than 1,000 circRNAs and 2,400 circRNA candidates in human serum and whole blood respectively (40).
Many efforts have been made to determine whether tumor-derived exo-circRNAs enter the circulation and are measurable for cancer detection. Human CDYL circRNA was readily detected in the serum of a tumor-bearing mouse xenograft model of human MHCC-LM3 cancer cells, and its abundance in the serum of xenografted mice was correlated with the tumor volume. Intriguingly, serum exosomal circRNAs were able to distinguish patients with colon cancer from healthy controls. Compared with healthy subjects, 67 circRNAs were missing and 257 new circRNA species were detected in CRC patients. Notably, many host genes for these new circRNAs (48 genes for 53 circRNAs) were significantly up-regulated in CRC tissue (38).
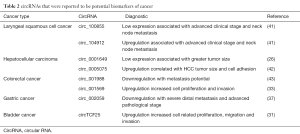
Many circRNAs have been found dysregulated in cancer, showing their potential as circulating biomarkers (Table 2). For instance, the expression of hsa_circ_001988 in CRC tissues was significantly down-regulated compared with normal mucosa, and related to tumor differentiation. In addition, the mean hsa_circ_001988 expression in perineural invasion positive tumors was lower than that in negative tumors (43). Another study revealed that hsa_circ_002059 was significantly down-regulated in gastric cancer, suggesting that lower expression of hsa_circ_002059 is associated with more severe distal metastasis and advanced cancer, and that hsa_circ_002059 may be a potential novel and stable biomarker for the diagnosis of gastric cancer (37). In hepatocellular carcinoma (HCC) tissues, hsa_circ_0001649 expression was significantly down-regulated. Hsa_circ_0001649 expression was lower in HCC tumors with a larger size suggesting that hsa_circ_0001649 exerted a suppressive effect on tumor growth (26). In laryngeal squamous cell cancer, hsa_circRNA_100855 was the most up-regulated circRNA and hsa_circRNA_104912 was the most down-regulated circRNA as compared with the corresponding adjacent non-neoplastic tissues. Patients with T3–4 stage, neck nodal metastasis or advanced clinical stage exhibited a higher hsa_circRNA_100855 expression and a lower hsa_circRNA_104912 expression (41). With more efforts devoted to this field, larger numbers of circRNAs will be disclosed. Highly sensitive RNA-seq analyses of normal and cancerous tissues identified a large number of circRNAs candidates, suggesting their potential biomarker roles. Based on a better understanding about the circRNA expression profile, further studies may focus on their sensitivity and specificity in cancer diagnosis and prognosis for clinical samples combined with a series of circRNAs or other cancer biomarkers.
Full table
Conclusions
CircRNA is a large group of competing endogenous RNAs that can affect the transcription and expression of genes. Dysfunction or change of circRNAs in cells is considered to be a sign of malignant degeneration, and a potential diagnostic or predictive biomarker for a variety of cancers as well. Nevertheless, the full picture of circRNAs in alimentary carcinogenesis remains unclear.
As one of the hot spots in cancer research, circRNAs are expected to become promising candidates for new methodology of RNA-based cancer therapy, knowing that their high cytoplasmic stability would make them long-acting regulators of cellular behavior. In view of their miRNA sponges function, remodeling of the circRNA-miRNA balance might be useful for cancer treatment, either by targeting the up-regulated miRNAs in cancer cells, or by blocking the miRNA-circRNA combination via saturating the conserved binding sites on circRNAs to enhance biological activity of miRNAs that is reduced in cancer. The therapeutic potentials of some circRNAs have been recently discovered. For example, circular miRNA sponged against miRNA-21, and miRNA-221displayed superior anti-cancer activities in melanoma cell lines (44).
The circRNA world is full of treasure compared with coding RNAs, miRNAs and lncRNAs. There are still significant gaps in our current understanding of circRNAs, such as circRNA degradation or turn-over and nomenclature standardization. With the development of technology and further research, increasing number of scientists will reveal functions of most circRNAs in tumor genesis and finally bring them from bench to bedside.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (81301498; 81271920); Jiangsu Provincial Program for Medical Innovation Teams and Leading Talents (LJ201133); Key Project of Jiangsu Province (BE2015654); Scientific Research Foundation for Jiangsu Provincial Commission of Health and Family Planning (H201526); Translational Medicine Project of Affiliated Hospital of Nantong University (TDF-zh201406).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.37). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Huang C, Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription 2015;6:61-4. [Crossref] [PubMed]
- Ghosal S, Das S, Sen R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 2013;4:283. [Crossref] [PubMed]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. [Crossref] [PubMed]
- Xin Z, Ma Q, Ren S, et al. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics 2017;16:80-6. [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA 2010;16:2043-50. [Crossref] [PubMed]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol 2010;20:R858-61. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016;13:34-42. [Crossref] [PubMed]
- Chao CW, Chan DC, Kuo A, et al. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 1998;4:614-28. [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Sand M, Bechara FG, Sand D, et al. Circular RNA expression in basal cell carcinoma. Epigenomics 2016;8:619-32. [Crossref] [PubMed]
- Qu S, Song W, Yang X, et al. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data 2015;5:385-7. [Crossref] [PubMed]
- Sand M, Bechara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 2016;83:210-8. [Crossref] [PubMed]
- Oncogenic Circular RNAs Arise from Chromosomal Translocations. Cancer Discov 2016;6:OF20.
- Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis 2012;33:1126-33. [Crossref] [PubMed]
- Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer 2011;47:1127-37. [Crossref] [PubMed]
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 2015;1-8. [PubMed]
- Wei F, Cao C, Xu X, et al. Diverse functions of miR-373 in cancer. J Transl Med 2015;13:162. [Crossref] [PubMed]
- Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016;16:161-9. [Crossref] [PubMed]
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015;6:6001-13. [Crossref] [PubMed]
- Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 2016;35:3919-31. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget 2016;7:63449-55. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. [Crossref] [PubMed]
- Horsham JL, Ganda C, Kalinowski FC, et al. MicroRNA-7: A miRNA with expanding roles in development and disease. Int J Biochem Cell Biol 2015;69:215-24. [Crossref] [PubMed]
- Chen YJ, Chien PH, Chen WS, et al. Hepatitis B Virus-Encoded X Protein Downregulates EGFR Expression via Inducing MicroRNA-7 in Hepatocellular Carcinoma Cells. Evid Based Complement Alternat Med 2013;2013:682380 [PubMed]
- Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015;29:2168-82. [Crossref] [PubMed]
- Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015;444:132-6. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015;61:221-30. [Crossref] [PubMed]
- Memczak S, Papavasileiou P, Peters O, et al. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One 2015;10:e0141214 [Crossref] [PubMed]
- Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 2016;8:932-9. [PubMed]
- Shang X, Li G, Liu H, et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine (Baltimore) 2016;95:e3811 [Crossref] [PubMed]
- Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol 2015;8:16020-5. [PubMed]
- Liu Y, Cui H, Wang W, et al. Construction of circular miRNA sponges targeting miR-21 or miR-221 and demonstration of their excellent anticancer effects on malignant melanoma cells. Int J Biochem Cell Biol 2013;45:2643-50. [Crossref] [PubMed]


