
MiRNA-516b inhibits ameloblastoma cell proliferation and invasion by regulating MYCBP/c-myc/RECK/MMP pathway
Introduction
Ameloblastoma (AM) is a frequent odontogenic tumor originating from dental epithelium. AM is characterized as invasively aggressive owing to its local invasiveness ability and higher risk of recurrence (1,2). AM accounts for more than 30% of all odontogenic tumors (3), which are typically classified as multicystic, peripheral, desmoplastic, unicystic, and ameloblastic carcinoma based on the World Health Organization classification of head and neck tumors (4). While surgical treatments are effective in preventing AM, severe facial deformity and psychological trauma are some of the consequences associated with the use of surgical treatments (5). Thus, identification of molecular aberrations is critical in order to find effective therapeutic strategies for the treatment of AM.
MicroRNAs (miRs) are small, non-coding endogenous RNAs (19–22 nucleotides) derived from double-strand RNA (6). miRs play important regulatory function by suppressing target gene expression by binding to the 3' untranslated region (UTR) of target mRNA, thereby resulting in the degradation of target mRNA or inhibiting translation (7). Many miRs are highly evolutionarily conserved among distant species, thus playing an important role in essential biological processes, such as cell proliferation, apoptosis, migration and signal transduction (6,8,9). Increasing evidence has showed that aberrant miRNAs expression exhibits oncogenic or tumor suppressive roles in human cancers (10-12). Diniz et al. has demonstrated that lower miR-15a/16-1 expression promotes the development of odontogenic tumor, thereby indicating an etiological and therapeutic role of miRs in odontogenic tumor (13). Recently, miR-516b has been reported to be significantly downregulated in several malignant tumors, including non-small cell lung cancer (NSCLC)(14), primary melanoma (15), and prolactinomas (16), thus suggesting its role in tumor suppression. However, to our knowledge, no studies have evaluated the expression and biological function of miR-516b in AM.
Thus, we pay an attention on the critical role of miR-516b in AM. We determined the expression levels of miR-516bin AM samples and normal cells. In this study, we investigated the role of miR-516b in AM and identified the underlying molecular mechanisms.
Methods
Tissue collection
Fresh AM tissues were obtained from AM patients, who received surgical treatment without any pre-operative chemotherapy or radiotherapy, at Sun Yat-sen Memorial Hospital, China. Normal oral mucosa was used as control. These tissues were frozen in liquid nitrogen and stored at −80 °C for the subsequent experiments. The study and the samples used were approved by the ethics committee of Sun Yat-sen Memorial Hospital. Written informed consent was obtained from all the patients before administering the clinical materials.
Cell culture and transfection
The human immortalized AM cell line, hTERT-AM and normal epithelial cells were obtained from Guanghua School and Hospital of Stomatology and Institute of Stomatological Research, Sun Yat-sen University (Guangzhou, China). The samples were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS, Gibco BRL, Grand Island, NY, USA) and 200 µg/mLG418 (Gibco, Grand Island, NY, USA) in a humidified atmosphere containing 5% CO2 at 37 °C.
For cell transfection, the cells were cultured in a complete medium without antibiotics for at least 24 hours. The cells were washed with 1× PBS (pH7.4) and transiently transfected with miR-516b mimic, mimic negative control (NC), MYCBP siRNA, and scramble siRNA (GenePharma, Shanghai, China) using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For MYCBP overexpression, the cells transfected with miR-516b mimic were plated in 24-well plates, pcDNA3.0-MYCBP and pcDNA3.0 blank vector (Invitrogen, USA) were transfected into the cultured cells, respectively, with Lipofectamine™ 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
RNA isolation and quantitative real time PCR (qRT-PCR)
Total RNA from tissues and cells was isolated using RNAiso (TaKaRa, Dalian, China) and reverse transcribed using PrimeScript RT reagent kit(TaKaRa) according to the manufacturer’s instructions. The mRNA expression levels were detected using the SYBR-Green PCR Master Mix kit (Takara) in PCR reaction. Reaction steps were as follows: 95 °C for 30 s, as the first step in a loop; 95 °C for 5 s, 60 °C for 34 s, as the second step, for 40 cycles. Then qRT-PCR was carried out using an ABI PRISM 7900HT Sequence Detection System (Thermo Fisher Scientific, Inc., MA, USA). The relative expression values of miR-516b was calculated and normalized to endogenous small nuclear RNA U6 control in each sample using the 2−ΔΔCt method. The primer sequences used in this study are as follows:
- Human miR-516b;
- RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAGTGCT;
- miR-516b: F: ACACTCCAGCTGGGATCTGGAGGTAAGAAGC;
- U6 F: CTCGCTTCGGCAGCACA;
- U6 R: AACGCTTCACGAATTTGCGT. Each sample was run in triplicates.
Cellular viability
After plating cells in 96-well microtiter plates (Corning Costar, Corning, NY, USA) at 1.0×103/well, cellular viability was determined after 24, 48 and 72 h using CCK-8 test (Dojindo, Kumamoto, Japan) by determining the absorbance of the converted dye at 450 nm.
Cell cycle and apoptosis analyses
Following cell transfection, the AM cells were seeded at a density of 3×105 cells per well in 6-cm dishes and collected until 80% confluence. For cell cycle analysis, the supernatant was discarded after centrifugation, and attached cells were harvested. The cells were fixed in cold 70% ethanol for 30 min at 4 °C and stained with propidium iodide (PI) solution (Sigma-Aldrich China, Inc., Beijing, China). Histograms of cell cycle distribution in G0/G, S, and G2/M phases were analyzed using a FACSort flow cytometer (Becton Dickinson, CA, USA) according to the manufacturer’s instructions. For apoptosis analysis, the cells were stained with Annexin V-APC/7-AADkit (BestBio, Shanghai, China). Stained cells were analyzed using FACSort flow cytometer (Becton Dickinson, CA)
Wound scratch assay
At 48 h post-transfection, the cells were incubated overnight until they had reached 85–95% confluence. Three scratches/well were made in each plate, using a 200 µL plastic filter tip. The detached cells were washed off using serum-free medium. The lodged cells were incubated in fresh complete medium at 37 °C in a 5% CO2 atmosphere. At 24 h post-scratching, the wound areas were photographed and observed using a microscope. The quantification of wound closure was performed using Zen2012 software by measuring the percentage of the wound area as compared to the total cell covered area. All scratch assays were performed in triplicates and the experiment was repeated thrice.
In vitro transwell invasion assay
The Transwell invasion assay was used assess the invasive ability of AM cells under different treatments. Briefly, the cells were cultured in serum-free media in the upper Transwell culture chambers, with 8-µm pore filter inserts pre-coated with BD Matrigel™ Basement Membrane Matrix (BD Biosciences, San Jose, CA, USA) overnight. The medium with 10% FBS was added to the lower chamber as chemoattractant. After 24 h of incubation, the cells attaching to the lower surface were fixed with 100% methanol, and stained with 0.1% crystal violet. The number of invasive cells on the lower chamber was counted in high-power fields (magnification, ×200), and the average invasive number was calculated as cell counts. All invasion assays were performed in triplicates and each independent experiment was repeated thrice.
Luciferase reporter assays
A dual luciferase assay was performed to confirm whether MYCBP directly bind to miR-516b. Briefly, the AM cells were reseeded in 24-well plates for 24 h and were transiently co-transfected with 0.3 µg MYCBP-3'-UTR-wild type or MYCBP-3'-UTR-mutant reporter plasmid (Agilent Technologies, USA), followed by transfection with corresponding 50 nM miR-51b mimic or miR-control using Lipofectamine 2000 (Invitrogen). After 48 h, Firefly and Renilla luciferase activities were analyzed using the Dual Luciferase Assay (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Each test was repeated thrice and each independent experiment was performed in triplicates.
Western blot analysis
Fresh AM tissue samples and cells were homogenized and lysed in a buffer containing 1X protease inhibitor cocktail (Thermo Fisher Scientific, Inc.). The protein concentration was determined using Bradford assay (Bio-Rad Laboratories, USA). Equivalent quantities of protein (30 µg) were separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel(10%) and transferred to a PVDF membrane. The membrane was blocked with 10% defatted milk and incubated overnight with primary antibodies targeting c-mycbp (Abcam, ab139534, 1:1000), c-myc (Abcam, ab10910, 1:000), RECK (Abcam, ab88249, 1:000), MMP2 (Abcam, ab14311, 1:000), MMP9 (Abcam, ab73734, 1:000), and GAPDH (Abcam, ab88249, 1:000), which are purchased from ABCAM (Cambridge, England). After washing with PBST thrice, the membrane was incubated with the corresponding secondary antibodies. The bound antibodies were detected using the enhanced chemiluminescence kit (Santa Cruz, USA).
Statistical analysis
Quantitative data is presented as the mean ± standard (SD) from three independent experiments. Significant differences in two independent groups were evaluated by t-test using SPSS 19.0 software. A value of P<0.05 was considered to be statistically significant.
Results
Regulation of MiR-516b in AM tissue specimens and cells
The expression levels of miR-516b were determined in all AM specimens and patient-matched normal specimens. The result showed that the expression of miR-516b was significantly downregulated in AM tissues as compared to the adjacent normal tissues (Figure 1A and Table 1, P<0.01). Additionally, the expression of miR-516b in hTERT-AM cell line was decreased as compared to the cells infected with sh-c-myc (Figure 1B and Table 2, P<0.001). These findings indicated that miR-516b plays an important role in the development of AM.
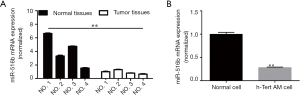
Full table

Full table
Effect of miR-516b on cell proliferation, cell migration and invasion, and cell cycle in AM
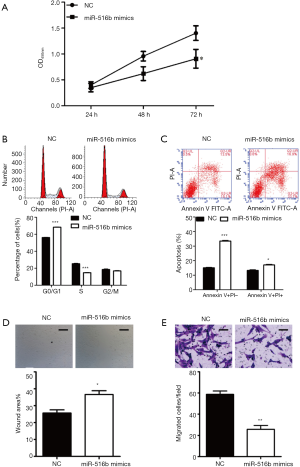
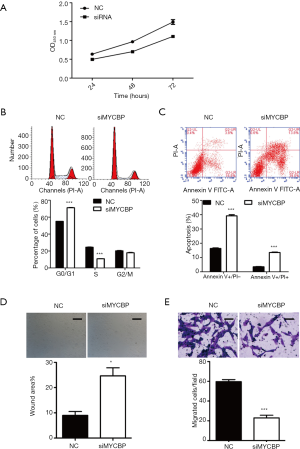
To investigate the biological effects of miR-516b in AM cells, the hTERT-AM cell line was first transfected with miR-516b mimic and mimic NC. Subsequently, a series of functional assays, including CCK-8, flow cytometry assay and Transwell assay, were used to identify AM cells transfected with miR-516b mimics. The CCK-8 assay showed that transfection with miR-516b mimic inhibited cell proliferation as compared to transfection with mimic NC (Figure 2A, P<0.05). Cell cycle analysis showed that the experimental group transfected with the miR-516b mimics displayed a significantly higher percentage of cells in the G0/G1 phase (68.55%±1.17% vs. 56.44%±0.97%) and a lower percentage of cells in the S phase (14.99%±0.76% vs. 25.91%±1.23%) as compared to the NC group (Figure 2B, P<0.001). Results from flow cytometry assay showed that miR-516b mimic induced early apoptosis and late apoptosis in AM cells, as indicated by increased cell percentage of early (Annexin V+/PI−) and late apoptosis (Annexin V+/PI+) as compared to NC group (Figure 2C, P<0.05, P<0.001). In addition, Wound scratch assay showed that wound areas in miR-516b mimic group was significantly decreased as compared to NC group, thereby suggesting impaired cell migration capacity induced by miR-516b overexpression (Figure 2D, P<0.05). Moreover, miR-516b was involved in the invasion of AM cells. As shown in Figure 2E, overexpression of miR-516b significantly decreased the invasion capacities of AM cells (P<0.01). Together, these results demonstrated that miR-516b suppresses tumor by suppressing proliferation and invasion in AM cells.
Role of MYCBP in AM cells
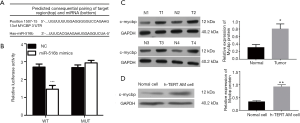
We identified MYCBP as a possible direct target gene of miR-516b, which is involved in the proliferation and migration of hTERT-AM cells, using predictive tools (TargetScan Human; http://www.targetscan.org/vert_71/) (Figure 3A). To verify the prediction, luciferase reporter assays were performed as described. The results showed that wild-type miR-516b significantly inhibited luciferase activity of MYCBP 3′UTR (Figure 3B, P<0.001), thereby indicting the direct regulation of miR-516b in the 3'UTR of MYCBP mRNA. Furthermore, we determined the expression of MYCBP in AM tissues and cells using Western blot analysis. As shown in Figure 3C and 3D, the expression of c-mycbp protein levels was significantly increased in AM tissues and cells as compared to control (P<0.05 or P<0.01). These results suggested that MYCBP could be an oncogene in AM, which is a direct target gene of miR-516b.
Effect of MYCBP on proliferation, migration and invasion, and cell cycle in AM cells
siMYCBP or scramble siRNA was transfected into hTERT-AM cells to investigate the role of MYCBP in AM cells by performing loss-of-functional experiments. As shown in Figure 4A, siMYCBP transfection significantly suppressed cell proliferation in AM cells as compared to scramble siRNA (P<0.01). Moreover, flow cytometry analysis showed that knockdown of MYCBP remarkably induced cell cycle arrest at G0/G1 phase (Figure 4B, P<0.001) and promoted early and late cell apoptosis (Figure 4C, P<0.001) in hTERT-AM cells. Similarly, it was also observed that both migration and invasion ability were significantly suppressed in siMYCBP group as compared to NC group in hTERT-AM cells, as demonstrated by Wound scratch assay (Figure 4D, P<0.05) and Transwell assay (Figure 4E, P<0.001), respectively. These findings indicated that downregulation of MYCBP could suppress cell proliferation and invasion of AM cells.
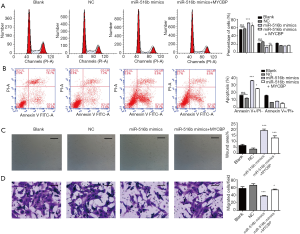
Effect of overexpression of MYCBP on AM cells transfected with miR-516b mimics in vitro
We determined whether upregulation of MYCBP could reverse the effects of miR-516b on the apoptosis, migration, and invasion of hTERT-AM cells. Forty-eight hours after pcDNA/MYCBP or pcDNA blank vector was transfected into miR-516b mimic, results showed that upregulation of MYCBP significantly weakened the effects of miR-516b overexpression on the G0/G phase arrest (Figure 5A) and induced early apoptosis (Figure 5B, P<0.05, P<0.001) of hTERT-AM cells. Additionally, Wound scratch assay (Figure 5C, P<0.001) and Transwell assay (Figure 5D, P<0.05, P<0.001) showed that overexpression of MYCBP could partially reverse the inhibition effects of miR-516 overexpression on cell migration and invasion in hTERT-AM cells.
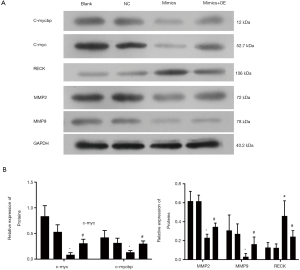
Regulation of c-myc/RECK/MMPs signaling pathway by MiR-516b
Protein levels of c-mycbp were determined. The level of miR-516b-overexpressionin AM cells was decreased as compared to control cells in a manner that was similar to c-mycbp expression induced by siMYCBP (Figure 6). Conversely, the protein levels of c-mycbp were dramatically increased when pcDNA/MYCBP was introduced into AM cells, which expressed miR-516b at a higher level (Figure 6, *, P<0.05 vs. NC group, #, P<0.05 vs. miR-516 mimics group). These results suggested that MYCBP could be a direct target and an important functional mediator of miR-516b. Further, we analyzed the expression of c-myc, RECK, and MMPs in AM cells. Similarly, miR-516b had a significant inhibitory effect on the expression of c-myc, MMP2, and MMP9, but significantly increased RECK expression in a way similar to siRNA-mediated knockdown of MYCBP (Figure 6, *, P<0.05 vs. NC group, #, P<0.05 vs. miR-516 mimics group). In addition, these effects were significantly rescued by MYCBP overexpression (Figure 6).
Discussion
Prognosis of AM in AM patients is poor due to its aggressive local invasiveness ability and high rate of recurrence (3). The molecular mechanisms involved in the development of AM are still not fully understood. MiRNAs have been reported to play a critical role in tumor initiation and progression through post-transcriptional regulation of gene expression (17). Previous studies have shown that miR-516b is expressed at low level in non-small cell lung cancer (NSCLC) (14), primary melanoma (15), and prolactinomas (16). However, little is known about the functional role and mechanistic action of miR-516b in AM. In the present study, we demonstrated that miR-516b was downregulated in AM tumor samples as compared to corresponding adjacent normal tissues. Also, we examined the effects of miR-516b on AM cells in vitro.
Our results indicated that overexpression of miR-516b significantly suppressed cell growth by inducing cell cycle arrest and apoptosis and inhibited cell migration and invasion capacity in AM cells, thereby suggesting tumor suppressive role of miR-516b in AM. However, the molecular mechanism of miR-516b in suppressing tumor cell proliferation remains unclear. Using luciferase reporter and Western blot analysis, we identified MYCBP as a potential direct target gene for miR-516b. We also found that c-mycbp was upregulated in AM tissues and cells. Subsequent studies have shown that inhibition of MYCBP significantly suppressed proliferation, migration, and invasion capacity of AM cells, and restoration of MYCBP partially rescued the inhibitory effect of miR-516b on AM cells. Therefore, we opined that miR-516b plays a critical role in the suppression of proliferation and metastasis of AM cells by downregulating the expression of c-mycbp.
MYCBP, initially identified as a MYC-interacting protein, acts as a positive modulator of mammalian Hedgehog (Hh) signaling pathway (18), which was previously demonstrated to affect the development of tumorigenesis (19,20). The activation of MYCBP through E-box-dependent transcription by c-myc contributes to tumor cell growth and invasion (19,20). In breast, tumor-suppressive miR-22 inhibits transcription of E-box-containing c-myc target genes by silencing c-mycbp (21). Together, these findings suggested that MYCBP could act as an oncogene in various cancers and facilitate the downregulation of miRNAs, including miR-516b in tumor. C-myc is considered to play a key role in inducing cell cycle progression, transformation, and apoptosis (22-25). These findings suggested that MYCBP is a direct target of miR-516b, thus making miR-516b a likely indirect regulator of c-myc. To test this assertion, we detected the expression of c-myc protein in AM cells after miR-516 overexpression or MYCBP depletion. The findings confirmed the hypothesis of indirect regulation of c-myc activity by miR-516b through modulating the expression of its cofactor, MYCBP.
RECK has been found to function as a tumor suppressor in many types of cancer (26-28). MMPs are extracellular matrix-degrading related enzymes that play a key role in tumor growth and invasion. MMP2 and MMP9 are the two important members of MMPs, which are inhibited by RECK at the post-transcriptional level (29,30). A recent study has showed that RECK is significantly downregulated in AM and its overexpression reduces invasive ability of AM cells by reducing the expression of MMP2 and MMP9 (31). In this study, it was observed that treatment with miR-516b mimic or siMYCBP could significantly inhibit c-myc expression in order to decrease the expression of MMP2 and MMP9. It was observed that RECK, MMP2, and MMP9 function as downstream effector proteins of MYCBP, which are indirectly regulated by miR-516b.
Conclusions
Our results suggest that miR-516b function as a potential target for tumor suppression by exerting its inhibitory effect on proliferation, migration, and invasion of AM cells by indirectly regulating c-myc/RECK/MMPs pathway by inducing MYCBP expression. However, further studies are required to validate its role in tumor suppression in AM.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of Guangdong (No.2014A030313025).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study and the samples used were approved by the ethics committee of Sun Yat-sen Memorial Hospital and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Faras F, Abo-Alhassan F, Israel Y, et al. Multi-recurrent invasive ameloblastoma: A surgical challenge. Int J Surg Case Rep 2017;30:43-5. [Crossref] [PubMed]
- Kumamoto H. Molecular pathology of odontogenic tumors. J Oral Pathol Med 2006;35:65-74. [Crossref] [PubMed]
- Fregnani ER, da Cruz Perez DE, de Almeida OP, et al. Clinicopathological study and treatment outcomes of 121 cases of ameloblastomas. Int J Oral Maxillofac Surg 2010;39:145-9. [Crossref] [PubMed]
- Ando T, Shrestha M, Nakamoto T, et al. A case of primordial odontogenic tumor: A new entity in the latest WHO classification (2017). Pathol Int 2017;67:365-9. [Crossref] [PubMed]
- Mendenhall WM, Werning JW, Fernandes R, et al. Ameloblastoma. Am J Clin Oncol 2007;30:645-8. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3' UTR in development and differentiation. Semin Cell Dev Biol 2005;16:49-58. [Crossref] [PubMed]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature Reviews Molecular Cell Biology 2008;9:219-30. [Crossref] [PubMed]
- Winter J, Diederichs S. MicroRNA biogenesis and cancer. Methods Mol Biol 2011;676:3-22. [Crossref] [PubMed]
- Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006;12:580-7. [Crossref] [PubMed]
- Jin J, Deng J, Wang F, et al. The expression and function of microRNA-203 in lung cancer. Tumour Biol 2013;34:349-57. [Crossref] [PubMed]
- Yu H, Lu Y, Li Z, et al. microRNA-133: expression, function and therapeutic potential in muscle diseases and cancer. Curr Drug Targets 2014;15:817-28. [Crossref] [PubMed]
- Diniz MG, Gomes CC, de Castro WH, et al. miR-15a/16-1 influences BCL2 expression in keratocystic odontogenic tumors. Cell Oncol (Dordr) 2012;35:285-91. [Crossref] [PubMed]
- Hu L, Ai J, Long H, et al. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget 2016;7:8441-54. [Crossref] [PubMed]
- Hanniford D, Segura MF, Zhong J, et al. Identification of metastasis-suppressive microRNAs in primary melanoma. J Natl Cancer Inst 2015;107. [PubMed]
- Wang C, Su Z, Sanai N, et al. microRNA expression profile and differentially-expressed genes in prolactinomas following bromocriptine treatment. Oncol Rep 2012;27:1312-20. [PubMed]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389-402. [Crossref] [PubMed]
- Lin C, Yao E, Wang K, et al. Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev 2014;28:2547-63. [Crossref] [PubMed]
- Taira T, Maëda J, Onishi T, et al. AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes to Cells 1998;3:549-65. [Crossref] [PubMed]
- Sakamuro D, Prendergast GC. New Myc-interacting proteins: a second Myc network emerges. Oncogene 1999;18:2942-54. [Crossref] [PubMed]
- Xiong J, Du Q, Liang Z. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene 2010;29:4980-8. [Crossref] [PubMed]
- Packham G, Cleveland JL. c-Myc and apoptosis. Biochim Biophys Acta 1995;1242:11-28. [PubMed]
- Henriksson M, Lüscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Advances in Cancer Research 1996;68:109-82. [Crossref] [PubMed]
- Hoffman B, Liebermann DA, Selvakumaran M, et al. Role of c-myc in myeloid differentiation, growth arrest and apoptosis. Curr Top Microbiol Immunol 1996;211:17-27. [Crossref] [PubMed]
- Ryan KM, Birnie GD. Myc oncogenes: the enigmatic family. Biochem J 1996;314:713-21. [Crossref] [PubMed]
- Xu M, Wang HF, Zhang HZ. Expression of RECK and MMPs in Hepatoblastoma and Neuroblastoma and Comparative Analysis on the Tumor Metastasis. Asian Pac J Cancer Prev 2015;16:4007-11. [Crossref] [PubMed]
- Wang L, Ge J, Ma T, et al. Promoter hypermethylation of the cysteine protease RECK may cause metastasis of osteosarcoma. Tumour Biol 2015;36:9511-6. [Crossref] [PubMed]
- Clark JC, Thomas DM, Choong PF, et al. RECK--a newly discovered inhibitor of metastasis with prognostic significance in multiple forms of cancer. Cancer Metastasis Rev 2007;26:675-83. [Crossref] [PubMed]
- Takahashi C, Sheng Z, Horan TP, et al. Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc Natl Acad Sci U S A 1998;95:13221-6. [Crossref] [PubMed]
- Oh J, Takahashi R, Kondo S, et al. The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 2001;107:789-800. [Crossref] [PubMed]
- Liang QX, Liang YC, Xu ZY, et al. RECK overexpression reduces invasive ability in ameloblastoma cells. J Oral Pathol Med 2014;43:613-8. [Crossref] [PubMed]


