
Monitoring response to therapy by testing circulating tumor DNA in urine
Husain and co-workers assessed the levels of epidermal growth factor receptor (EGFR) mutations in urine samples of advanced non-small cell lung cancer (NSCLC) patients receiving the third generation EGFR tyrosine kinase inhibitor (TKI) osimertinib (1). This drug has shown to be active in NSCLC patients with sensitizing EGFR mutations (such as deletions in exon 19 and the p.L858R point mutation in exon 21) who become resistant to first- or second-generation EGFR TKIs through the development of the p.T790M EGFR resistance mutation (2). The study enrolled nine EGFR-mutant advanced NSCLC patients who progressed on first- or second-generation TKIs and had confirmed both a sensitizing EGFR mutation and the p.T790M variant in their NSCLC tissue biopsy. The p.T790M mutation was detected in the pre-treatment urine samples of eight patients. Analysis of serial urine samples for EGFR mutations showed a pattern of intermittent spikes throughout the first week of treatment and an overall decrease in fragment numbers at day 7. This dynamic of EGFR mutations in urine suggests drug-induced tumor apoptosis within days of initial treatment, followed by a decrease associated to tumor response. In this respect, the decrease of EGFR mutations in urine preceded the radiographic response assessed at 6 to 12 weeks of treatment.
These findings add novel information in a field that is rapidly expanding, i.e. the use of liquid biopsy in the management of cancer patients. The term liquid biopsy refers to the possibility to perform tumor molecular profiling by using tumor-derived biomarkers that can be isolated from body fluids of cancer patients, including peripheral blood, urine, cerebrospinal fluid, effusions. In cancer patients’ fluids different types of biomarkers (DNA, RNA, proteins, miRNA) can be isolated from different sources (biomarkers can be free or within cells, exosomes, platelets etc.) (3). However, the main field of application of liquid biopsy is currently represented by the analysis of circulating cell free DNA (cfDNA), which contains both DNA deriving from normal cells as well as the circulating tumor DNA (ctDNA) that is released by tumor cells through different mechanisms.
The majority of available data on liquid biopsy relate to the use of plasma as source of cfDNA, although increasing evidence suggests that urine might represent a valuable alternative to plasma to isolate tumor-derived DNA (4,5). Indeed, urine-based testing is a truly non-invasive procedure that may potentially facilitate both diagnostic and monitoring capabilities without requiring visits to the clinic (Table 1). The collection of samples is easy and it may therefore save time and procedure costs compared to both tissue biopsy and plasma-based testing. However, we must acknowledge that limited information is available on the pre-analytic and analytic factors that might affect urine-based cfDNA testing, including the best time of the day for urine collection, the stability of tumor-derived nucleic acids in the urine and the most adequate procedure for DNA extraction. Furthermore, no data are available on the correlation of urine EGFR mutations levels and response to therapy.
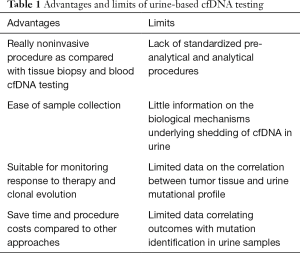
Full table
The results of Husain and collaborators are in line with previous reports suggesting the potential to monitor response to therapy by using plasma-based cfDNA testing. However, discordant data have been described with respect to the presence of spikes of mutant DNA following treatment with different drugs. Marchetti et al. did not find early spikes of EGFR mutations in cfDNA obtained from plasma samples of EGFR-mutant metastatic NSCLC patients treated with EGFR TKIs (6). Serial samples were analyzed starting 4 days after treatment up to 60 days. Patients showing early reduction of EGFR mutations in cfDNA were more likely to respond to therapy. Similarly, Tie and co-workers did not find significant difference in plasma mutation levels between pretreatment and three days after chemotherapy in 53 metastatic colorectal carcinoma patients (7). However, the authors found that 3 out of 4 patients, who had a spike in mutant DNA 3 days after chemotherapy followed by a rapid decline, had an excellent response. In melanoma patients receiving tumor infiltrating immunotherapy the presence of an early serum peak of the BRAF V600E mutation was strongly associated with the likelihood to obtain an objective response (8).
The discordant results reported in these studies might be due to different factors. First, the number of patients analyzed in most of these reports is extremely limited, thus preventing from the possibility to drive any firm conclusion. It is also possible that plasma and urine cfDNA have different kinetics. Indeed, Riediger et al. have described a case of a patients with an EGFR mutant NSCLC in whom a peak of EGFR mutation in plasma cfDNA was detected 24 to 48 hours after treatment with an EGFR TKI (9). Because in other studies plasma sampling started 4 days after treatment, this approach might have missed earlier changes in mutation load. In this regard, biological and anatomical mechanisms might lead to a later peak in the urine as compared with plasma. Finally, the kinetic of induction of plasma spikes might depend on pharmacologic properties of the employed drugs. In this respect, serial analysis of plasma EGFR mutation levels in a recent case report of osimertinib in a patient with acquired T790M-mediated resistance to gefitinib also showed an increase after three days followed by a rapid clearance (10). The reduction in mutation load was associated with significant response to treatment.
The analysis of genetic alterations in liquid biopsy requires high sensitive techniques, due to the dilution of ctDNA in normal DNA. In this respect, Husain and colleagues used a deep sequencing approach leading up to a 0.006% limit of detection (LOD) (1). An increasing number of laboratories are using next generation sequencing (NGS) approaches to analyze cfDNA (11). However, the majority of NGS panels available on the market have a limited sensitivity. New NGS technologies with enhanced sensitivity are becoming available (3). In addition to the low LOD, the main advantage of NGS is the ability to interrogate a high number of loci per analysis, being therefore able to provide information on known and novel genetic alterations of different type (single nucleotide variants, indels, copy number variation, gene fusions). Indeed, NGS analysis of liquid biopsy has revealed that resistance to EGFR TKIs is often multi-clonal rather than being monoclonal (12). This information might be clinically relevant, because patients with multiple mechanisms of resistance are likely to have a suboptimal response to treatment with agents targeting a single pathway of resistance (2). Therefore, NGS will probably replace in short time the current methods for mutation testing in clinical research and clinical diagnostics (13).
In conclusion, the data reported by Husain and colleagues open novel, interesting scenarios for the use of liquid biopsy in cancer. The possibility to perform daily sampling of cfDNA through urines might allow early assessment of response to any kind of systemic therapy, either targeted agents or standard chemotherapy or immunotherapy. The introduction in the clinical scenario of an early marker of response or resistance will favor the development of novel therapeutic strategies in which the treatment will be adjusted based on the predicted sensitivity to the drug and on the presence of molecular mechanisms of resistance that can be detected in the liquid biopsy. In this respect, the advancement in liquid biopsy is essential for the further development of precision medicine in oncology.
Acknowledgments
Funding: This work was supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to N. Normanno (Grant number: IG17135).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shaohua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.43). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Husain H, Melnikova VO, Kosco K, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin Cancer Res 2017;23:4716-23. [Crossref] [PubMed]
- Normanno N, Maiello MR, Chicchinelli N, et al. Targeting the EGFR T790M mutation in non-small-cell lung cancer. Expert Opin Ther Targets 2017;21:159-65. [Crossref] [PubMed]
- Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Fujii T, Barzi A, Sartore-Bianchi A, et al. Mutation-Enrichment Next-Generation Sequencing for Quantitative Detection of KRAS Mutations in Urine Cell-Free DNA from Patients with Advanced Cancers. Clin Cancer Res 2017;23:3657-66. [Crossref] [PubMed]
- Marchetti A, Palma JF, Felicioni L, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 2015;10:1437-43. [Crossref] [PubMed]
- Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015;26:1715-22. [Crossref] [PubMed]
- Xi L, Pham TH, Payabyab EC, et al. Circulating Tumor DNA as an Early Indicator of Response to T-cell Transfer Immunotherapy in Metastatic Melanoma. Clin Cancer Res 2016;22:5480-6. [Crossref] [PubMed]
- Riediger AL, Dietz S, Schirmer U, et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep 2016;6:33505. [Crossref] [PubMed]
- Vallee A, Audigier-Valette C, Herbreteau G, et al. Rapid clearance of circulating tumor DNA during treatment with AZD9291 of a lung cancer patient presenting the resistance EGFR T790M mutation. Lung Cancer 2016;91:73-4. [Crossref] [PubMed]
- Deans ZC, Williams H, Dequeker EMC, et al. Review of the implementation of plasma ctDNA testing on behalf of IQN Path ASBL: a perspective from an EQA providers' survey. Virchows Arch 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Normanno N, Rachiglio AM, Roma C, et al. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. Journal of cellular biochemistry 2013;114:514-24. [Crossref] [PubMed]


