
Microphthalmia-associated transcription factors activate mTORC1 through RagD GTPase gene expression
The mechanistic target of rapamycin complex 1 (mTORC1) is a major serine/threonine kinase that stimulates cellular anabolic processes including protein and lipid synthesis while suppressing catabolic processes such as autophagy in response to growth factors and amino acids (1). Upon mTORC1 activation, it phosphorylates multiple substrates including S6 kinase (S6K), eIF4E binding protein (4EBP), and unc-51-like kinase (ULK) (2). Both S6K and 4EBP are key regulators for mRNA translation and cell cycle progression (3). In addition to mTORC1’s roles in stimulating these anabolic processes, mTORC1-dependent ULK phosphorylation inhibits its kinase activity, which is essential for autophagy induction (4). Thus, mTORC1 activation in response to growth factors and amino acids promotes key cellular anabolic processes while it suppresses major catabolic processes, to build biosynthetic molecules essential for cell growth and proliferation.
Both growth factor and amino acid signals impinge on the lysosomal membrane and coordinately stimulate the activity of mTORC1 by enhancing two distinct lysosomal small GTPases, Rheb and Rags, respectively. While the Rags recruit mTORC1 to the lysosomal membrane in response to amino acids such as leucine, arginine, and glutamine (5), Rheb, which directly interacts with mTORC1, stimulates the activity of mTORC1 on the lysosomal membrane in response to growth factors (6). Mammalian cells contain four members of Rag small GTPases (RagA, B, C, and D) and form obligate heterodimers of either RagA or RagB with either RagC or RagD (7). In the active Rag heterodimer, RagA or RagB binds to GTP while RagC or RagD binds to GDP. Upon amino acid availability, the Ragulator complex, a guanine nucleotide exchange factor (GEF) for RagA/B (8,9), stimulates RagA/B GTP loading in a manner dependent of lysosomal vATPase activity (10). Likewise the folliculin (FLCN)-FLIP complex, a GTPase activating protein (GAP) stimulates RagC/D GDP loading (11). Once the Rag heterodimer is in its active configuration, it localizes mTORC1 to the lysosomal membrane (5). On the other hand, growth factors instigate the activation of the PI3K (phosphoinositide 3-kinase)/Akt pathway leading to the activation of Rheb small GTPase, which directly stimulates mTORC1 on the lysosome (6).
It is not surprising that nutritional signals collide on the lysosomal membrane. The lysosome is responsible for breaking down macromolecular components through the process of autophagy. Autophagy allows cells to respond to stress conditions such as starvation by providing nutrients through the degradation of cellular components (12). Once nutrients become available, mTORC1 is activated, stimulating anabolism while ending the autophagy response. Thus the lysosome serves as a sensing platform where the extracellular and intracellular nutritional status is carefully monitored in order to maintain a balance between catabolic and anabolic processes.
mTORC1 is well known to inhibit the induction of autophagy by phosphorylation of ULK (4). In addition, mTORC1 has also been shown to negatively regulate two transcription factors, transcription factor EB (TFEB) and transcription factor E3 (TFE3) (13-15), which play a key role in inducing the expression of numerous genes encoding lysosomal hydrolases, membrane proteins, and essential proteins for autophagy. Both TFEB and TFE3 are members of the microphthalmia-associated transcription factor (MiTF) subfamily of transcription factors (16). Upon mTORC1 activation, it phosphorylates these transcription factors at key serine residues, which creates a binding site for the 14-3-3 cytosolic chaperone protein, leading to the blockade of nuclear translocation of these transcription factors (17). In contrast, under starvation conditions, mTORC1 is inactivated, thus the dephosphorylated form of TFEB and TFE3 dissociates from its interaction with the 14-3-3 protein and localizes to the nucleus. Here TFEB and TFE3 recognize the coordinated lysosomal expression and regulation (CLEAR) elements in the promoter region of genes responsible for lysosomal biogenesis and autophagy (16,18). Consequently, TFEB and TFE3 transcriptionally up-regulate the capacity of degradation machineries in cells to generate nutrients for their survival under starved conditions.
Previously Martina et al. reported that TFE3 could function as part of a feedback loop leading to mTORC1 activation by increasing expression of the FLCN and two FLCN interacting proteins FNIP and FNIP2, which form the FLCN-FNIP complex, a GAP that activates RagC/D small GTPases (19). Consistently, TFE3 overexpression stimulates Rag C/D GDP loading necessary for its activation, leading to lysosomal mTORC1 localization and its activation. The same observation was also made in a model of TFEB overexpression. Thus, continuous TFE3 and TFEB activation prepare the source and machinery for mTORC1 activation and might ensure the termination of autophagy-lysosomal-mediated catabolism once nutrients become available.
Understanding the molecular mechanisms by which TFEB/TFE3 regulate mTORC1 activity is particularly relevant as the mutations in TFEB/TFE3 genes have been found in renal cell carcinoma and amongst other cancers with high mTORC1 activity (16,17). Elucidation of the molecular mechanisms by which TFEB/TFE3 mutations lead to aberrant mTORC1 activation would provide insight into the development of possible therapeutic strategies.
In this context, the study recently published in Science by Di Malta et al. provided crucial roles of MiTF transcription factor family members, substrates of mTORC1, in the activation of mTORC1. Interestingly, the study indicates that even under amino acid sufficient conditions, the inhibition of TFEB or TFE3 leads to a decrease in cellular mTORC1 activity in a variety of mammalian cells. These results suggest that the transcription factors TFEB and TFE3 responsible for the initiation of lysosome biogenesis and autophagy under starvation conditions are indeed required for mTORC1 activity in response to amino acids.
Yu et al. have previously shown that upon amino acid starvation, the activity of mTORC1 is abolished as expected, however, prolonged amino acid starvation restores cellular mTORC1 activity (20). They proposed that mTORC1 is stimulated in response to the availability of newly synthesized nutrients restored by autophagy during prolonged starvation conditions, bringing to an end the autophagy response. In support of this model, genetic ablation of ATG5 or ATG7, key proteins for autophagy, inhibited the restoration of mTORC1 activity under prolonged starvation conditions (20).
The study by Di Malta et al., also observed that the restoration of mTORC1 activity in response to prolonged starvation was abolished when TFEB/TFE3 were genetically ablated, indicating that these transcription factors are crucial for mTORC1 re-activation under this condition. It could be argued that the loss of mTORC1 re-stimulation under prolonged starvation conditions is due to a decrease in the capacity of cellular autophagy-lysosome degradation system caused by the lack of TFEB/TFE3-dependent expression of lysosomal and autophagic proteins. However, the authors showed that TFEB overexpression lead to higher mTORC1 activity in cells lacking the essential autophagy genes, ATG5 or ATG7 compared to control cells. Based on these results the authors propose that the MIT-TFE transcription factors may stimulate mTORC1 activity in a manner independent of their role in the induction of autophagic machinery.
Leucine and arginine have been shown to be two amino acids particularly important for mTORC1 activation on the lysosomal membrane (21,22). The study demonstrated that the sensitivity of mTORC1 activation in response to leucine or arginine was increased in cells overexpressing TFEB, yet the complete starvation of leucine largely inhibited mTORC1 activity in these cells. These results suggested that TFEB/TFE3 overexpression might support the expression of positive regulators in amino acid sensing machinery responsible mTORC1 activation. Likely candidates include the FLCN complex and the subunits of v-ATPase, which is a positive regulator of the Ragulator complex that functions as a GEF for the RagA/B small GTPases.
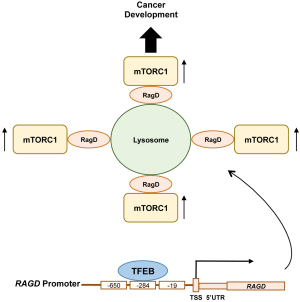
As expected, enhanced gene expression in the TFEB overexpressing cells includes previously known TFEB targets important for amino acid-induced mTORC1 activation. Interestingly, the authors also identified RAGD as a putative TFEB target gene that encompasses the CLEAR element in its promoter. Strikingly, among 20 TFEB/TFE3 putative target genes that likely involve in the regulation of mTORC1 activity, RAGD was the most decreased transcript in TFEB or TFE3 silenced cells, whereas it was the most enhanced gene in cells overexpressing TFEB. Of note, consistent with the previous report by Martina et al. (19), FLCN expression was likewise affected by TFEB expression but to a much lesser extent to that compared of RAGD. Through chromatin immunoprecipitation (CHIP) and luciferase assays, the authors confirmed that RAGD is a direct transcriptional target of TFEB, as the RAGD promoter has three CLEAR sites upstream of its transcriptional start site (Figure 1). Functional importance of the RAGD CLEAR element was confirmed by generating cells bearing a deletion of a key endogenous CLEAR site through CRISPR-CAS9-mediated genome editing (RagDpromedit cells). In RagDpromedit cells, amino acid-induced lysosomal mTORC1 localization and its activation were significantly decreased compared to the control cells that have the intact TFEB binding CLEAR element. These observations indicate that TFEB/TFE3-induced RAGD expression plays an important role in amino acid-induced lysosomal mTORC1 localization and its activation. Although the data indicate an important role of endogenous RagD in the activation of mTORC1, it remains unclear why transcriptional inhibition of the RAGD gene is so effective for mTORC1 inhibition in the presence of RagC of which expression is more ubiquitous and has a redundant function with RagD for mTORC1 regulation in response to amino acid availability. It is possible that the Rag heterodimer containing RagD might have higher activity and/or additional roles for lysosomal mTORC1 recruitment compared to the RagC containing heterodimer.
Based on these observations made in an in vitro system, the authors also addressed whether the over expression of TFEB or TFE3 had physiological relevance in tissues particularly important in adaptation to nutrient and starvation signals. In a liver specific TFEB over expression model, mTORC1 activity was indeed enhanced under nutrient rich conditions, but was inhibited under fasting conditions. In contrast, muscle specific TFEB knockout mice showed decreased mTORC1 activity in response to a post exercise leucine oral gavage, which was used to emulate the effect of a protein meal after exercise. These results pinpoint that TFEB is necessary to mediate leucine-mediated mTORC1 stimulation in vivo. Although their in vitro studies showed that TFEB is required for mTORC1 activation in response to leucine or full amino acid stimulation, it remains elusive if lack of TFEB also blocks full amino acid-induced mTORC1 activation in muscle tissues (23). In addition, the investigation of RagD expression and its role in mTORC1 activation in response to amino acid feeding in the exercised muscles will further clarify physiological relevance of the TFEB-RagD axis in the regulation of mTORC1 activity in vivo.
The TFEB/TFE3/MITF transcription factors belong to the MiT-TFE transcription factor family, and are well known oncogenes in various human tumors including renal cell carcinoma, melanoma, sarcoma and pancreatic ductal adenocarcinoma, in which aberrant mTORC1 activation is also observed. The study demonstrated positive correlations among the expression of TFE3/MITF, RagD expression, mTORC1 activity, cancer cell proliferation, and tumor development. Consistent with the other biochemical and biological observations demonstrated in this study, renal cancer cells carrying a chromosomal translocation of the TFE3 gene, pancreatic ductal adenocarcinoma bearing high MiT/TFE genes, and melanoma cells with aberrant MITF expression all showed increased RAGD transcript accompanied with increased mTORC1 activity. Silencing either these transcription factors or RagD attenuated mTORC1 activity as well as cell proliferation in these cancer cells, implying that the MiT/TFE-RagD-mTORC1 axis plays an important role in cancer cell proliferation/survival in vitro. Importantly, xenotransplantation experiments performed using the melanoma cell line showed significant reduction of xenografted tumor development upon RAGD silencing, highlighting a critical role of the MiT-TFE-RagD axis in promoting tumor development.
The MiT/TFE transcription factors are active in their dephosphorylated form under starvation conditions when mTORC1 is inactivated. However, the study reported by Di Malta et al. proposed a model where MiT/TFE-RagD-mTORC1-MiT/TFE feedback circuit is crucial for metabolic adaptation to nutrient availability. Dys-regulation of this circuit such as constitutive activation of MiT/TFE leads to aberrant RagD-mediated mTORC1 activation and promotes cancer development (Figure 1). One likely physiological role of this feedback circuit is that under metabolic stress conditions, these transcription factors stimulate RagD expression and would prepare lysosomal mTORC1 localization and its activation once nutrients are replenished through extracellular influx or de novo production by autophagy. Alternatively, the MiT/TFE-RagD axis may play an emergent and specific role in keeping a low level of mTORC1 activity, maintaining cellular translational activity for the transcripts of lysosomal and autophagy components, as the restored mTORC1 activity after prolonged starvation is required for lysosomal biogenesis (20). In this regard, it is intriguing to examine the specific role of inducible RagD in the activation of mTORC1 under metabolic stress conditions.
In conclusion, this study provided a novel molecular mechanism by which oncogenic MiT/TFE transcription factors support cell growth/proliferation through their transcriptional regulation of the upstream of mTORC1 activator, RagD. The MiT/TFE-RagD-mTORC1-MiT/TFE feedback circuit precisely controls anabolic and catabolic processes with appropriate checkpoints and balances to maintain cellular homeostasis (Figure 1). Upon MiT/TFE overexpression as that observed in a variety cancers, a loss of anabolic/catabolic homeostasis occurs, leading to increased cell growth and proliferation even under metabolically stress conditions by enhancing RagD expression, and thus increased mTORC1 activity.
Acknowledgments
Funding: This work is supported by grants from the National Institute of Health (NIH DK083491, GM110019, T32GM008322) and the Department of Defense (DOD TS140055).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Zhongnan Hospital of Wuhan University, Wuhan, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.31). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009;122:3589-94. [Crossref] [PubMed]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006;124:471-84. [Crossref] [PubMed]
- Fingar DC, Richardson CJ, Tee AR, et al. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol 2004;24:200-16. [Crossref] [PubMed]
- Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132-41. [Crossref] [PubMed]
- Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496-501. [Crossref] [PubMed]
- Inoki K, Li Y, Xu T, et al. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003;17:1829-34. [Crossref] [PubMed]
- Yao Y, Jones E, Inoki K. Lysosomal regulation of mTORC1 by amino acids in mammalian cells. Biomolecules 2017;7. [PubMed]
- Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010;141:290-303. [Crossref] [PubMed]
- Bar-Peeled L, Schweitzer LD, Zoncu R, et al. Ragulator is a GEF for the Rag GTPases that signal smino scid levels to mTORC1. Cell 2012;150:1196-208. [Crossref] [PubMed]
- Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011;334:678-83. [Crossref] [PubMed]
- Tsun ZY, Bar-Peled L, Chantranupong L, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013;52:495-505. [Crossref] [PubMed]
- Lim CY, Zoncu R. The lysosome as a command-and-control center for cellular metabolism. J Cell Biol 2016;214:653-64. [Crossref] [PubMed]
- Martina JA, Chen Y, Gucek M, et al. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012;8:903-14. [Crossref] [PubMed]
- Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012;31:1095-108. [Crossref] [PubMed]
- Roczniak-Ferguson A, Petit CS, Froehlich F, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012;5:ra42. [Crossref] [PubMed]
- Settembre C, Fraldi A, Medina DL, et al. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013;14:283-96. [Crossref] [PubMed]
- Slade L, Pulinilkunnil T. The MiTF/TFE family of transcription factors: master regulators of organelle signaling, metabolism and stress adaptation. Mol Cancer Res 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473-7. [PubMed]
- Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014;7:ra9. [Crossref] [PubMed]
- Yu L, McPhee CK, Zheng L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010;465:942-6. [Crossref] [PubMed]
- Wolfson RL, Chantranupong L, Saxton RA, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016;351:43-8. [Crossref] [PubMed]
- Chantranupong L, Scaria SM, Saxton RA, et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016;165:153-64. [Crossref] [PubMed]
- Lee JH, Cho US, Karin M. Sestrin regulation of TORC1: is Sestrin a leucine sensor? Sci Signal 2016;9:re5. [Crossref] [PubMed]


