
Liquid biopsies: envisioning a future when tissue is avoidable in lung cancer treatment decision-making
During the last years we have witnessed a revolution in the genomic profiling and molecular characterization of lung cancer. Epidermal growth factor receptor (EGFR) is, among other signaling pathways such as ALK, BRAF, MET or ROS, one of the key oncogenes with potential for targeted-inhibition. Therefore, a complete basal genomic profiling is now recommended in the clinical diagnostic workup to select the optimal personalized therapy for each lung cancer patient (1).
Treatment with first- and second-generation EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib, gefitinib and afatinib, are currently considered the standard of care at first-line in patients with sensitive mutations (Exon 19 Deletions, L858R) at the kinase domain of the EGFR gene (1). Despite the initial outstanding responses, patients ultimately progress after a median of 9–12 months. One of the most common mechanism of resistance described is the threonine-to-methionine substitution at amino acid position 790 (T790M) in exon 20 of the EGFR gene that occurs in approximately 50–60% of patients. Other less frequent bypass track mechanisms have also been reported such as the activation of MET, HER2, AXL, IGF1R, or FGFR1 (2).
Osimertinib (AZD9291) is an oral, selective third-generation, irreversible EGFR TKI with activity against both EGFR-TKIs sensitizing and T790M resistance mutations (3). In November 2015 and April 2016, the US Food and Drug Administration (FDA) (4) and the European Medicines Agency (EMA) (5) respectively, approved the use of osimertinib at the recommended 80 mg daily dose, in advanced non-small cell lung cancer (NSCLC) patients harboring a T790M mutation whose disease progressed on EGFR-TKI therapy, based on the data of the expansion cohort of the phase I study (namely AURA) with osimertinib and two phase II studies (AURA extension and AURA 2). The results reinforced the efficacy of osimertinib, in T790M-mediated resistant patients, with remarkable overall response rate (ORR) (66–71%), encouraging progression-free survival (PFS) (9.7–11 months) and a manageable safety profile (6).
The confirmatory trial was a randomized, phase III trial (AURA3) in TKI-resistant, EGFR T790M-positive and advanced NSCLC patients. Osimertinib significantly improved PFS (10.1 vs. 4.4 months; hazard ratio 0.30; P<0.001) and ORR (71% vs. 31%; P<0.001) with better toxicity profile (adverse events ≥ grade 3: 23% vs. 47%) and QoL as compared with standard platinum-pemetrexed chemotherapy (7).
However, all this innovation in terms of effective treatments and new molecular knowledge in the field of EGFR NSCLC represents a clinical challenge because at disease progression, a new tumor re-biopsy is endorsed in order to identify the underlying mechanism of resistance (1). In turn, this has become a major matter of concern as small biopsies or cytological specimens are the only source of material in almost 80% of NSCLC patients (8) and tumor sampling often requires invasive procedures that might be associated with a higher risk of complications and an increase in the average cost per patient (9). Moreover, genetic results from a single tumor-biopsy do not mirror tumor heterogeneity or allow for monitoring molecular resistance changes along time (10).
In the last years circulating-free tumor DNA (ctDNA), also known as “liquid biopsy”, has emerged as a sensitive and feasible, non-invasive and cost-effective alternative to tissue biopsies to screening for genetic drivers in advanced NSCLC patients. New platforms using blood-based differ in terms of sensitivity, specificity, turnaround time and the rate of mutation detection. Amplification refractory mutation system (ARMS), Cobas and peptide-nucleic acid (PNA) are non-digital polymerase chain reaction (PCR)-based platforms with semi-quantitative ability for mutant ctDNA analysis. Newer platforms, including droplet digital PCR (ddPCR) and BEAMING (beads, emulsion, amplification and magnetics) PCR, allow to quantify the amount of mutant ctDNA, whilst next generation sequencing (NGS) allow for a more extensive analysis of a range of genomic alterations (11,12).
Updated ESMO and NCCN guidelines (1,13) have incorporated ctDNA testing in their algorithms as an available and complementary technique for T790M mutation detection in EGFR-mutant NSCLC at disease progression. Among the different blood-based diagnostic tests, the only one that has recently granted approval by the FDA to aid clinicians in selecting patients with metastatic NSCLC with specific sensitive (exon 19 and 21) and resistant (T790M) EGFR mutations is the Cobas EGFR mutation testing platform v2 (Cobas® EGFR test) (14).
The study published recently by Remon et al. in Annals of Oncology (15), aimed to prospectively assess the efficacy of osimertinib in a cohort of TKI-resistant EGFR-mutant NSCLC patients when T790M status was determined solely and exclusively in ctDNA from blood samples by using an Inivata InvisionTM enhanced Tagged Amplicon-Sequencing technology (eTAm-SeqTM) (16).
The study included 48 EGFR-mutant advanced NSCLC patients from a single institution between April 2015 and April 2016. Patients had to have advanced NSCLC, a common sensitizing EGFR-mutation (Del19, L858R), clinical or radiological progression to at least one first- or second-generation EGFR TKI and importantly be ineligible for a new tissue re-biopsy for T790M resistance screening. Upon response evaluation criteria in solid tumors (RECIST) progression, those patients T790M-positive, by means of ctDNA-blood testing, were treated with osimertinib (80 mg daily).
Primary objective was to determine the ORR with osimertinib by RECIST on the basis of a positive T790M mutational status from ctDNA. Secondary endpoints included PFS (by radiological or investigator’s criteria) as well as the percentage of T790M positive identified in blood ctDNA.
In the evaluated cohort, ctDNA T790M mutation was detected in 50% (24/48) NSCLC patients. For the primary analysis, among assessable patients (n=16), osimertinib gave an ORR of 62.5%. With a median follow-up of 8.5 months, median PFS was not achieved (95% CI: 4–not available) and was 66.7% and 52% at 6- and 12-months respectively.
ctDNA T790M testing in NSCLC: clinical validation of the Cobas assay
The aforementioned health agencies approval of the PCR-based Cobas assay for T790M plasma genotyping in EGFR-mutant NSCLC was prompted by the results of the retrospective analysis from paired blood and tissue samples collected from patients included in pivotal trials of osimertinib (AURA phase I–II trials).
A first assessment using Cobas and BEAMing platforms was made in a cohort of patients (n=72) included in the AURA phase 1 study of osimertinib. The two platforms demonstrated moderate sensitivity (73–81%) and specificity (67–58%) respectively. The ORR in patients positive for plasma T790M was almost identical to that of patients positive for T790M mutation in paired-tissue samples (59% vs. 61%) (17). In a second analysis of the same phase I AURA trial, both plasma and tissue genotyping for T790M mutation by BEAMing were analyzed in 216 patients. Sensitivity and specificity of T790M-positive detection by ctDNA was 70% and 69% respectively. Of 58 patients with T790M-negative by tumor tissue 31% (n=18) were T790M-positive by plasma. Of these 18 discordant cases (considered “false positives”), 14 (78%) were confirmed using an alternative plasma assays suggesting a real T790M mutation. Both, ORR (63% and 64%) and median PFS (9.7 and 7.9 months) were almost identical in T790M-positive patients detected in plasma and tissue respectively. Interestingly, better outcomes than expected were observed in plasma T790M-negative patients with an ORR of 46% and median PFS of 8.2 months. Among plasma T790M-negative subgroup those with tumor T790M-positive, obtained an ORR of 69% and median PFS of 16.5 months (similar results to overall plasma T790M-positive population) (18).
A larger cohort of 511 patients with matched plasma and tissue samples were evaluated in the phase II trials (AURA extension and AURA2). In these studies, Cobas EGFR mutation plasma test v2 genotyping platform was employed and compared with both Cobas tumor tissue genotyping and NGS plasma genotyping platforms. ORR in patients with T790M-positive by both Cobas tissue and plasma platform was similar (64% and 66% respectively). ORR for patients evaluated by plasma NGS was not reported. Sensitivity and specificity for Cobas plasma compared with tissue genotyping was 61% and 79% and compared with orthogonal NGS plasma genotyping assay 93% and 92% respectively. A total of 27 patients considered T790M-positive by plasma were T790M-negative by tissue test. These discordant cases were assessed further by plasma NGS and 23 (85%) of them were confirmed as positive too (19).
Outcomes of ctDNA T790M-positive patients of pivotal AURA 3 cohort have been recently reported. Cobas plasma T790M genotyping obtained a sensitivity and specificity of 51% and 77% respectively compared with tissue genotyping with the same platfform. Within the ITT populations, patients T790M-positive by plasma and randomized to treatment (n=179) obtained a median PFS of 8.2 months and a ORR 77% similar to obtained by tumor positive T790M population (median PFS 10.1 months and ORR 71%) (20).
However it is salient to mention that all these plasma genotyping validation studies, with the exception of the Oxnard’s cohort (18), were done exclusively in those patients with tissue positive T790M status, so this could have biased the results reported with regards the value of plasma in capturing those tissue T790M false negative cases.
ctDNA T790M testing in NSCLC: the hidden pitfalls
In addition to Cobas, a multitude of other platforms have reported validation reports for ctDNA T790M mutation detection in blood (Table 1). By using tissue genotyping as “reference”, digital and non-digital PCR platforms exhibit a consistent moderate sensitivity (51–81%) and specificity (58–79%). Albeit, prospective data is only available for a limited number of these studies, while others report data based on very small number of cases.

Full table
Due to the high positive predictive value of plasma of these platforms and the high rate of false negative cases, authors recommended that plasma genotyping should be used as a complementary tool for T790M testing. The true percentage of T790M false-positive cases detected by plasma genotyping is probably lower than reported because some of them are false negative cases of tumor genotyping. If plasma genotype is positive for T790M this result predicts excellent outcome but if you get a negative T790M plasma result guidelines recommend to perform a re-biopsy for tissue genotyping to discard a true false negative plasma genotyping and to rule out other mechanisms of resistance such as HER2, MET amplification or small cell lung cancer transformation (1,14).
Various degrees of validation exist for individual plasma genotyping assays and the need of using tissue-genotyping as the reference standard method for the majority of validated genotyping assays to date is still a matter of debate. On the one hand, T790M mutation might be as well underestimated in tissue biopsies due to spatial and temporal tumor heterogeneity (10) and on the other side new orthogonal NGS genotyping assays might be more sensitive (90–95%) (Table 1) than the aforementioned tests and capture a broader spectrum of genetic features in blood (22,23).
The study published by Remon et al. (15), differs from the others because they prospectively evaluated osimertinib efficacy but using only ctDNA for T790M-mutation detection without a matched tumor tissue sample used as reference. As for the platform used, eTam-SeqTM, it is a deep sequencing assay that allows for more extensive multiplexing including hotspot regions and copy number variations with potential predictive roles for patient’s outcome. They detected three different cases of concomitant mutations (PI3KA, STK11 and NRAS) along with T790M mutation. Indeed, this NGS platform obtained high sensitivity rates (92–100%) in preliminary analytical validation (25); in line with other NGS test (22,23). The T790M-positive rate of 50% and the similar outcomes obtained with osimertinib (ORR 62.5% and 6 months-PFS 66.7%) are consistent with previous studies in the same patient population (17-20). However, consideration should be given to the small sample size of patients evaluated for outcome (n=16) calling into question the reproducibility of the results. Moreover, as results were focused solely on T790M-positive population, data about false negative, sensitivity or specificity cannot be extrapolated. Similar to other studies (19) in this cohort no correlation was found between RECIST response and the frequency of T790M allelic fraction (AF). One explanation could be the long interval of time between the blood test and the start of osimertinib (median time of 1.5 months). Indeed, turnaround time of plasma NGS assays is potentially slower than other PCR-based assays such as Cobas. Correlations between RECIST response with osimertinib and ctDNA predictors (T790M AF, EGFR activating mutation AF and ratio of T790M and EGFR activating mutation AF) were also evaluated, and a non-significant trend was observed for lower mutant AFs of T790M or EGFR sensitizing mutations. The overall burden of metastatic disease in a patient has been previously demonstrated to predict for increased plasma genotyping sensitivity (24). However, in Remon’s study, no data about tumor volume was reported and no correlation was given between tumor burden and depth of response challenging the real significance of low AF detection in this plasma samples.
ctDNA T790M testing in NSCLC: looking ahead
Based on the concept that ctDNA T790M-positive detection is a good predictor of osimertinib treatment outcome it is reasonable to move forward designing prospective trials in which treatment decisions are based on plasma genomic testing results. The APPLE trial (NCT02856893) is a phase II trial for EGFR-mutant TKI-naïve NSCLC patients to evaluate the best strategy of treatment sequencing with gefitinib and Osimertinib (26). All patients (n=159) will be randomized (1:1:1): arm A will receive osimertinib as first-line treatment until disease progression according to RECIST criteria; whereas arms B and C will explore the procedure to switching from gefitinib to osimertinib at disease progression. However, and of key interest, definition of progression varies between later arms: arm B will employ molecular progression, that is to say plasma ctDNA detection of T790M-positive mutation, while arm C will use conventional RECIST disease progression criteria (Figure 1). This trial is based on the use of liquid biopsy by Cobas EGFR Mutation Test V2 in front of classical radiological criteria as a tool for making treatment decisions. They hypothesize that anticipating osimertinib use to RECIST progression by using plasma T790M testing will impact positively in patients’ outcome (26).
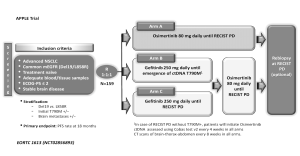
However, based on the recent outstanding results reported with osimertinib in the first line setting (AURA phase 1 trial) with median PFS of 22 months (27), the role of liquid biopsy for T790M detection is definitively uncertain. The use of other blood-platforms that allows for a more extensive multiplex analysis, such as NGS, might be more appropriate to identify other new mechanisms of resistance to osimertinib.
Conclusions
Current guidelines use ctDNA as a complementary tool to detect T790M as a mechanism of resistance at EGFR-TKIs progression. Multiple new platforms with different technical features have recently entered in the landscape of NSCLC and have obtained different results in terms of sensitivity and specificity being Cobas EGFR mutation testing platform v2, the first blood test approved in the clinic for guiding treatment decisions in NSCLC.
Remon et al. have reported the first evidence and “proof of principle” of the potential of liquid biopsies in the real-world setting, by electing personalized-treatment strategies based solely on plasma ctDNA instead of the standard tissue-genotyping. However, and for the time being, it is advised to carefully validate beforehand all these platforms against the ‘standard’ albeit ‘non-optimal’ tissue-genotyping in the setting of prospective clinical trials before their routine use in clinical practice.
Prospective clinical trials comparing higher sensitive platforms such as NGS for T790M detection in tissue, plasma or other biological liquids at disease progression are needed to establish the real role and future of ctDNA in daily clinical practice.
Acknowledgments
We want to thank Dr. J. Remon for his useful comments on the APPLE trial design.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Wei Xu (Jiangsu Provincial Key Laboratory of Geriatrics, Department of Geriatrics, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.20). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-v27. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools) - Summary of product characteristics. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/004124/WC500202022.pdf
- Yang J, Ramalingam SS, Jänne PA, et al. LBA2_PR: Osimertinib (AZD9291) in pre-treated pts with T790M-positive advanced NSCLC: updated Phase 1 (P1) and pooled Phase 2 (P2) results. J Thorac Oncol 2016;11:S152-3. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Bubendorf L, Lantuejoul S, de Langen AJ, et al. Nonsmall cell lung carcinoma: diagnostic difficulties in small biopsies and cytological specimens: Number 2 in the Series "Pathology for the clinician" Edited by Peter Dorfmüller and Alberto Cavazza. Eur Respir Rev 2017;26.
- Lokhandwala T, Bittoni MA, Dann RA, et al. Costs of Diagnostic Assessment for Lung Cancer: A Medicare Claims Analysis. Clin Lung Cancer 2017;18:e27-e34. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Sacher AG, Komatsubara KM, Oxnard GR. Application of Plasma Genotyping Technologies in Non-Small Cell Lung Cancer: A Practical Review. J Thorac Oncol 2017;12:1344-56. [Crossref] [PubMed]
- Jovelet C, Madic J, Remon J, et al. Crystal digital droplet PCR for detection and quantification of circulating EGFR sensitizing and resistance mutations in advanced non-small cell lung cancer. PLoS One 2017;12:e0183319 [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- US Food and Drug Administration. Cobas EGFR Mutation Test v2. Available online: https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm504540.htm
- Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017;28:784-90. [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68 [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Wu YL, Jenkins S, Ramalingam S, et al. MA08.03 Osimertinib vs Platinum-Pemetrexed for T790M-Mutation Positive Advanced NSCLC (AURA3): Plasma ctDNA Analysis. J Thorac Oncol 2017;12:S386.
- Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin Cancer Res 2016;22:2386-95. [Crossref] [PubMed]
- Li BT, Janku F, Janne PA, et al. Ultra-deep next generation sequencing (NGS) of plasma cell-free DNA (cfDNA) from patients with advanced lung cancers: results from the Actionable Genome Consortium. AACR 2016: Abstracts 2697-5293. New Orleans, Philadelphia: CTI Meeting Technology 2016:4342.
- Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Gale D, Plagnol V, Lawson A, et al. Analytical performance and validation of an enhanced TAm-Seq circulating tumor DNA sequencing assay. AACR 107th Annual Meeting 2016. New Orleans Cancer Res 2016;76:3639. [Crossref]
- Remon J, Menis J, Hasan B, et al. The APPLE Trial: Feasibility and Activity of AZD9291 (Osimertinib) Treatment on Positive PLasma T790M in EGFR-mutant NSCLC Patients. EORTC 1613. Clin Lung Cancer 2017;18:583-8. [Crossref] [PubMed]
- Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2017; [Epub ahead of print]. [Crossref] [PubMed]


