
Physiologically-based pharmacokinetic modeling as an approach to evaluate the effect of covariates and drug-drug interactions on variability in epidermal growth factor receptor kinase inhibitor exposure
Introduction
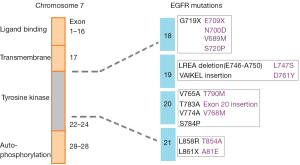
Epidermal growth factor receptor (EGFR) small molecule kinase inhibitors (KIs) have made a significant improvement in the treatment of advanced non-small cell lung cancer (NSCLC). Currently, there are four KIs approved for the treatment of NCSLC, with afatinib, erlotinib, and gefitinib used as first-line agents for the treatment of EGFR activating mutation positive tumors, and osimertinib is active against acquired T790M mutation mediated resistance after such treatment (1). The use of EGFR KIs for the treatment of this cancer is particularly interesting and has become a prominent example of personalised medicine where treatment is determined based on the mutation status of the individual’s tumour (2). EGFR activating mutations occur at a high frequency in NSCLC with 10% to 50% of tumours being positive for a mutation. It has been established that the treatment effect of EGFR inhibitors is superior for tumours harbouring an activating mutation compared to those without (3,4). Common mutations that confer either sensitivity or resistance to EGFR KIs are shown in Figure 1 and include alterations at exon 18, 19, 20 and 21 (5).
Tumour heterogeneity, environmental, demographic, and genetic factors can affect exposure to, and the therapeutic efficacy of, afatinib, erlotinib and gefitinib often resulting in highly variable clinical responses to these drugs in patients (6,7). In particular, inter-individual variability in drug exposure has been widely reported in a number of studies where a large proportion of patients experience either a lack of efficacy due to sub-therapeutic dosing or toxicity as a result of excessive dosing (8,9). It is well established that afatinib, erlotinib and gefitinib are extensively metabolised by cytochrome P450 (CYP) 3A4. Wide inter- and intra- individual variability in the activity of this enzyme due to factors effecting expression is widely reported. In addition, these EGFR KIs are also substrates for various transporters, organic anion transport peptide (OATP) 1B1 and p-glycoprotein (p-gp), in the intestinal tract and liver, which are all known to impact on exposure (10-13).
As NSCLC is responsible for the highest number of cancer related deaths globally, EGFR KIs are frequently prescribed to terminally ill patients at high doses. In addition to the many physiological factors, that influence exposure to EGFR KIs, given the environment that they are prescribed in, it is very likely that patients are co-administered other medications. Many commonly prescribed medications such as antibiotics (e.g., clarithromycin) and antifungals (e.g., fluconazole) are also metabolised by CYP3A4 and are known to alter the activity of this enzyme, and consequently may alter EGFR KI exposure. Given that patients receive a standard fixed dosing regimen, it is no surprise that significant inter-individual variability is an issue. Although many clinicians recognise this, and research toward optimal EGFR KI dosing has received significant attention, the solution is still unclear and currently unaddressed in clinical practice due to a lack of prospective evidence. Therefore, to assist in optimised EGFR KI dosing, the pharmacokinetics and hence exposure to these drugs is a key research focus.
Physiologically-based pharmacokinetic (PBPK) modeling, is a mechanistic “bottom up” approach, whereby the concentration-time profile for a drug in a particular patient cohort is simulated based on the physiochemical and in vitro kinetics of the drug and the physiological characteristics of the patient cohort. PBPK modeling is routinely utilised for the prediction of pharmacokinetic behaviour of new chemical entities during drug development, where it can be applied to investigate the potential impact of covariates such as age, gender, and metabolic drug-drug interactions (DDIs) on drug exposure in various population groups (14-16). While, it has been recognised that CYP3A4 is the major enzyme responsible for the metabolic clearance of afatinib, erlotinib and gefitinib, the exact contribution of CYP3A4 to the clearance of these drugs is less clear (9). EGFR KIs exhibit complex pharmacokinetic behaviour with permeability and or solubility limited absorption, variable distribution profiles, complex metabolism, typically resulting in the formation of multiple metabolites and substantial transporter mediated clearance resulting in enterohepatic recirculation.
In this study, we assessed the in vitro human liver microsomal metabolism of afatinib, erlotinib and gefitinib using a substrate depletion approach that allowed for the total CYP, total uridine diphosphate (UDP)-glucuronosyltransferase (UGT), and specific CYP3A4 mediated metabolism to be quantified without the necessity to individually quantify the formation of each metabolite. The in vitro intrinsic clearance for afatinib, erlotinib and gefitinib determined by substrate depletion were used to develop PBPK modeling profiles for each compound. These profiles were validated using data from healthy volunteer trials. The capacity to predict the impact of covariates such as age, gender, and ethnicity reported in clinical trials, and the impact of co-administration with strong CYP3A4 inhibitors and inducers on in vivo clearance were assessed using the Simcyp simulator (version 15.1).
Methods
Chemicals and reagents
Afatinib, erlotinib and gefitinib were purchased from Selleckchem (Boston, MA). Alamethicin, gucose-6-phosphate (G-6-P), G-6-P dehydrogenase, nicotinamide adenine dinucleotide phosphate (NADP), NADPH reductase, and UDP-glucuronic acid (UDP-GlcUA) were purchased from Sigma-Aldrich (St Louis, MO). Acetonitrile, ammonium acetate, formic acid and methanol were purchased from Merck Millipore (Melbourne, Australia). High purity water was obtained using a MilliQ Synergy UV Ultrapure water system (Merck Millipore, Sydney, Australia). All other solvents and reagents were of analytical grade or higher.
Enzyme and substrate preparations
Human liver microsomes (HLMs) were pooled from five individual livers (H7, H10, H12, H13, H40) obtained from the human liver bank of the Department of Clinical Pharmacology, Flinders University (Adelaide, Australia). Microsomes were prepared according to the method of Bowalgaha et al. [2005] (17). Approval was obtained from the Southern Adelaide Clinical Research Ethics Committee for the use of human liver tissue in drug disposition studies in vitro. HLMs were activated by pre-incubation with alamethicin (50 mg/mg microsomal protein), as described by Boase and Miners [2002] (18). HLM protein content was assessed according to standard measures (19). Afatinib, erlotinib and gefitinib were prepared in methanol such that the final solvent concentration in microsomal incubations was 1%.
Quantification of substrate depletion
The rate of microsomal KI metabolism was quantified in incubation samples (2,000 µL) containing HLM (0.5 mg/mL), phosphate buffer (0.1 M; pH 7.4) and substrate (1 µM) in the presence and absence of CYP (1 mM NADPH generating system) and/or UGT (5 mM UDP-GlcUA) cofactors (20,21). The contribution of non-CYP3A4 metabolism was quantified in the presence of the selective CYP3A4 inhibitor (CYP3Cide; 0.5 µM). Following a 10-min pre-incubation, reactions were initiated by the addition of substrate and the rate of depletion was assessed over 3 hours. Reactions were terminated with 4% acetic acid in methanol and samples were kept on ice. The supernatant fraction was isolated by centrifugation (5,000 g, 10 min, 10 °C). Substrate concentrations quantified in aliquots (200 µL) collected at 0, 30, 45, 60, 90, 120, 150, 180, 240, 300 and 360 min were used to calculate the rate of KI clearance for incubations performed in the presence of the appropriate cofactors and inhibitor (CYP3A4).
Substrate concentrations were quantified by high performance liquid chromatography (HPLC) (Agilent 1100 series instrument; Agilent Technologies, Sydney, Australia) with UV detection at the λmax for each analyte. Analytes were separated on a Waters NovaPak C18 analytical column [150 mm × 3.9 mm (id), 5 µm particle size; Waters Corporation, Milford, MA] using 10 mM ammonium acetate (pH 5.7; Mobile Phase A) with a gradient of 10% to 50% acetonitrile (Mobile Phase B) over 5 min.
Data analysis
All experiments were performed in triplicate and the mean analyte concentration was calculated from the integrated peaks in the chromatograms obtained from the HPLC. Total CYP and total UGT hepatic clearance was calculated based on the depletion half-life for incubations performed in the presence of the associated cofactors, using the equation:
Where CLint is the in vitro intrinsic clearance, Vinc is the incubation volume, t1/2 is the substrate half-life in the incubation, P is the amount of protein in the incubation and fu is the fraction of substrate unbound in the incubation (22). CYP3A4 mediated metabolism was calculated by subtracting the CLint determined in the presence of CYP3Cide from total CYP CLint. Microsomal CLint were used as model inputs defining the metabolic clearance for each drug in the Simcyp profiles.
PBPK structural model
Simulations were conducted utilising the Simcyp Simulator® (version 15.1) (23). Absorption was simulated using the advanced dissolution, absorption and metabolism (ADAM) sub-model, which considers various compartments of the gastrointestinal tract and several processes such as dissolution, gastrointestinal fluid transit, gut wall permeation, drug degradation, intestinal metabolism, and active transport (24). The ADAM absorption sub-model was used in conjunction with the whole body PBPK “fully-PBPK” model, comprising individual organ compartments (25-27). The differential equations used by the simulator describing enzyme kinetics and the impact of co-variates have been described previously (15).
PBPK population profile
EGFR KI profiles were developed and validated utilising the Simcyp Healthy Volunteer population comprising of 100 healthy individuals divided across ten trials with ten subjects each. Virtual subjects were aged between 20–50 years with a 50:50 female to male ratio. For simulations performed in cancer patients, the Genentech Cancer Population (28) was used; this population comprised patients aged 26–50 years with a 50:50 female to male ratio.
PBPK compound profiles
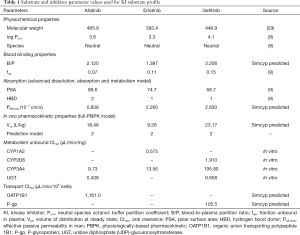
PBPK profiles for each compound were created based on individual reported physicochemical and distribution characteristics (9,29) and in vitro microsomal CLint data assessed by substrate depletion. In the absence of robust in vitro data describing intestinal and hepatic transporter kinetics, these parameters were estimated using the Simcyp parameter estimation function. Pharmacokinetic data used to create the compound profile for each EGFR KI are summarised in Table 1. Once developed, profiles for each compound were validated by comparing pharmacokinetic parameters [area under the curve (AUC), Cmax and tmax)] describing the simulated concentration-time profiles with those observed in clinical studies that were not used in the development of the profiles. Simulated and observed pharmacokinetic parameters for each EGFR KI are summarised in Table 2.
Full table

Full table
Assessing the effect of various covariates on exposure
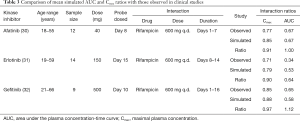
The effect of gender on EGFR KI exposure assessed by comparing exposure in all male and all female cohorts to an age and ethnicity matched cohort with a 50:50 female to male distribution. The effect of age on EGFR KI exposure was assessed by comparing exposure in a geriatric population aged 65–90 years to a healthy 20–50 years old population. The effect of ethnicity was also assessed in Japanese, Chinese and South African population comprising males and females (50% female) aged 20–50 years. Simulations were also performed in various disease models. The impact of covariates on EGFR KI exposure are summarised in Table 3.
Full table
Results
Substrate depletion by HLM
No depletion of afatinib, erlotinib and gefitinib by HLM was observed in the absence of co-factor. For incubations containing UDP-GlcUA, <10% substrate depletion was observed over the 3-hour incubation (Figure 2). When NADPH was present in the absence of CYP3Cide, substrate depletion of up to 90% was observed over 3 hours. The substrate depletion in incubations performed in the presence of both UDP-GlcUA and NADPH was equivalent to the sum of the depletion observed in incubation with each cofactor individually. Minimal (<10%) substrate depletion was observed in the presence of CYP3Cide with NADPH. Intrinsic clearance values adjusted for non-specific binding to incubation components are summarised in Table 1. Observed clearances in the presence and absence of CYP3Cide were consistent with a major contribution of CYP3A4 to the metabolism of these drugs (Figure 3).

Validation of EGFR KI profiles
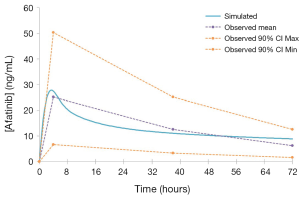
A comparison of mean [±95% confidence interval (CI)] model simulated pharmacokinetic parameters describing EGFR KI exposure with those observed in clinical studies are summarised in Table 2, and visualised in Figure 4. All ratios of observed to simulated pharmacokinetic parameters were contained within the range 0.8 to 1.2, and in all cases simulated pharmacokinetic parameters describing EGFR KI exposure were contained within the 95% CI for the observed parameter. A comparison of parameter (AUC and Cmax) ratios describing the impact of induction on EGFR KI exposure are summarised in Table 3. With the exception of the ratio of the AUC ratios for erlotinib (0.64), all observed to simulated parameter ratios describing the change in exposure caused by induction of clearance were contained within the range 0.8 to 1.2. As for parameters describing EGFR KI exposure, all simulated parameter ratios describing the impact of induction on EGFR KI exposure were contained within the 95% CI for the observed ratio.
Impact of covariates on simulated EGFR KI exposure
The influence of age, disease, dose, ethnicity and gender on simulated exposure to EGFR KIs was assessed by comparison of the mean AUC in covariate populations to a “control” population (n=50 participants) of healthy 20–50 years old Caucasians (50% female). Simulated populations (n=50 participants) were matched to the control group for all covariates except the one tested.
The mean AUC and % change from the control population for each population are reported for afatinib, erlotinib and gefitinib in Tables 4–6, respectively. In terms of physiological covariates, gender and renal function had no significant effect on afatinib, erlotinib or gefitinib exposure. Older age was associated with an increase in exposure for all EGFR KIs; mean AUCs were 20.5% (P=0.075), 30.4% (P=0.000) and 40.9% (P=0.001) higher in the geriatric (65–90 years old) population for afatinib, erlotinib and gefitinib, respectively. Morbid obesity was associated with a significant decrease in AUC for afatinib (−29.8%) and erlotinib (−26.2%) but not for gefitinib. Asian ethnicity was associated with significant increases in EGFR KI exposure; in the Chinese population AUC was significantly increased by 47.4% and 56.0% (P=0.000) for erlotinib and gefitinib, respectively, while afatinib did not significantly affect AUC. Erlotinib AUC significantly increased by 32.1% (P=0.004) while afatinib and gefitinib showed insignificant increases in the Japanese population. In contrast, the South African (mixed race) population had no effect (<10%) in AUC for afatinib, erlotinib or gefitinib. Liver dysfunction (Child Pugh A to C), had no significant change for afatinib AUC, however, erlotinib and gefitinib demonstrated significant changes in AUC (45–173%; P<0.005) with more severe liver dysfunction (Child Pugh B and C). No changes in AUC were predicted in the cancer population. When considering the impact of metabolic DDIs, administration of the CYP3A4 inducer rifampicin (600 mg q.d.) for 7 days prior to EGFR KI dosing caused decreases in AUC of 33.7%, 69.5% and 46.5% for afatinib, erlotinib and gefitinib, respectively. Co-administration of the CYP3A4 inhibitor ketoconazole (400 mg q.d.) increased afatinib, erlotinib and gefitinib AUCs by 28.8%, 103.0% and 77.3%, respectively.
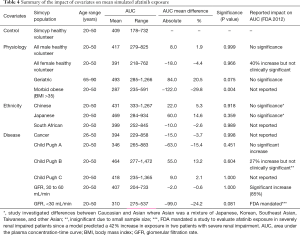
Full table
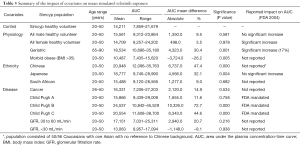
Full table
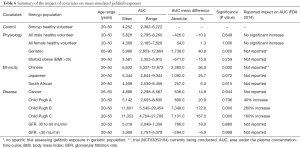
Full table
Application to optimised EGFR KI dosing
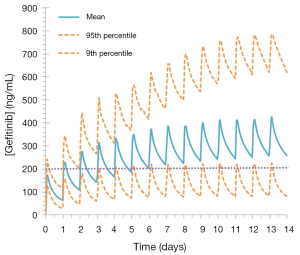
The established target threshold trough concentration for gefitinib that are associated with optimal therapeutic outcomes is >200 ng/mL (33). Simulated mean (range) trough gefitinib (250 mg q.d.) concentrations in 1,000 cancer patients following dosing to steady state (14 days) were 259.0 (range, 57.2–784.9) ng/mL. In this virtual population, 45% of patients treated with gefitinib had trough concentrations of <200 ng/mL at day 15 (Figure 5).
Discussion
Here we report for the first time a collection of sophisticated full-PBPK models for the first-line EGFR KIs used in the treatment of NSCLC. Although outcomes for patients with NSCLC have improved notably since the introduction of EGFR KIs, it is increasingly accepted that outcomes for NSCLC patients may be further enhanced by optimising exposure to these drugs (12,34,35). Variability in EGFR KI exposure primarily involves processes that occur in the gastrointestinal tract and liver, but can also be influenced by the tissue distribution of these drugs. In addition, EGFR KIs are known to be Biopharmaceutical Classification System (BCS) class II or IV drugs, where oral absorption can be complicated by permeability and/or solubility. Therefore, in order to simulate exposure to these drugs, profiles based on full-PBPK structural models with ADAM absorption sub-models that considered the compartmental distribution and interaction between permeability limited diffusion, transport and metabolism were developed using the Simcyp simulator®.
The key difference between the full-PBPK profiles developed here and previously reported minimal EGFR KI PBPK profiles (36-38) is that these full-PBPK profiles accurately account for the full range of physiochemical, metabolic and transporter medicated processes that determine exposure to these drugs. By way of example, in the current profiles, by accounting for permeability limited absorption through the use of the ADAM sub-model, these profiles accurately simulated not only the parameters defining EGFR KI absorption, but also the “shape” of the absorption profile. In contrast, previous models have either arbitrarily modified the absorption rate constant to shift or applied a non-physiological “lag time” to artificially model tmax.
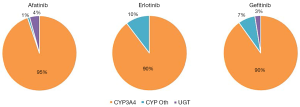
In the current study, substrate depletion was used to assess the contribution of enzyme families to and kinetics of, human liver microsomal EGFR KI metabolism (22). This approach was chosen as it facilitated the assessment of the enzymatic contribution to microsomal EGFR KI metabolism at physiologically relevant EGFR KI concentrations, and it allowed for the determination of microsomal EGFR KI clearances without the need to individually quantify the substantial number of EGFR KI metabolites. Control experiments performed in the absence of UDP-GluUA or NADPH demonstrated essentially no (<2%) depletion of afatinib, erlotinib, and gefitinib by HLM over the 3-hour incubation period. Similarly, less than 10% depletion was observed when incubations were performed in the presence of UDP-GlcUA alone (Figure 2), suggesting that UGT enzymes do not play a major role in the hepatic metabolism of these EGFR KIs. This observation is consistent with published literature regarding the contribution of UGT to the metabolism of these drugs (9). When NADPH was present in the absence of CYP3Cide, substrate depletion of up to 95% was observed over the 3-hour incubation period; this observation is consistent with the reported major contribution of CYP enzymes to the hepatic clearance of these drugs. The rate of substrate depletion observed in incubations containing UDP-GlcUA and NADPH was equivalent to the sum of the rates observed in incubations performed in the presence of each cofactor separately. This observation demonstrates that CYP and UGT catalyzed pathways have negligible effect on each other. The specific contribution of CYP3A4 to EGFR KI depletion was assessed by subtracting the depletion observed in the presence of NADPH with CYP3Cide (i.e., non-CYP3A4 oxidative metabolism) from the depletion observed in the presence of NADPH only (CYP3A4 and non-CYP3A4 metabolism). Less than 10% depletion was observed when CYP3Cide was present in incubations, indicating that for these EGFR KIs, CYP3A4 accounts for >90% of in vitro metabolism at substrate concentrations <1 µM. Calculated unbound microsomal CLint values for CYP3A4 catalysed metabolism were 9.73, 13.65 and 195.800 µL/min/mg for afatinib, erlotinib, and gefitinib, respectively. Remaining microsomal CLint values are reported in Table 1. These in vitro substrate depletion data were used to develop the compound profiles for each of the EGFR KI.
The EGFR KI compound profiles were validated by comparing simulated pharmacokinetic parameters (AUC, Cmax and tmax) describing exposure with those observed in clinical studies that were not used in the development of the compound profiles. With the exception of the AUC ratio describing the impact of induction on erlotinib exposure (0.64), the ratio of observed to simulated parameters describing exposure, or parameter ratios describing the impact of induction on exposure were contained within the range 0.8 to 1.2. This consistent and high degree of concordance demonstrates the accuracy and validity of the profiles, which is visually depicted in the representative overlay of the simulated and population average concentration-time curves defining afatinib exposure (Figure 4).
A major challenge for this study related to the validation of the capacity of the compound profiles to account for the impact of induction on EGFR KI exposure. As EGFR KIs exhibit substantial inter-individual variability in exposure and studies used to assess the impact of drug interactions are often undertaken in small (n<10) cohorts, the absolute exposure reported in these studies typically poorly reflects the population average. These studies are however adequately powered to accurately reflect changes in exposure (i.e., intra-individual variability) due to DDIs. As such, when validating the compound profiles for induction and inhibition, parameter ratios (i.e., the change in exposure) were considered rather than absolute exposures pre- and post-interaction.
Understanding and developing approaches to account for the covariates that impact on EGFR KI exposure is an important aspect of optimising the dosing of these drugs. In terms of physiological covariates, older age (i.e., geriatric population) consistently resulted in higher exposure to EGFR KIs, while morbid obesity resulted in moderately reduced exposure to erlotinib and afatinib, but not gefitinib. In contrast, gender did not affect afatinib, erlotinib, and gefitinib AUC.
Substantial increases in simulated EGFR KI exposure were also observed in age and gender matched Asian (Chinese and Japanese) populations. These simulations indicate that ethnicity may be an important factor in determining exposure to EGFR KIs. The clinical implications in terms of EGFR KI efficacy and tolerability and the physiological differences between Caucasian and Asian individuals that underpin these differences in exposure warrant investigation. Consistent with the mechanism of KI clearance, increasing severity of liver dysfunction, but not renal dysfunction, was associated with a marked increase in EGFR KI exposure. Most notably, erlotinib and gefitinib exposure was increased 2- to 3-fold in virtual patients with moderate or severe (Child Pugh B or C) hepatic dysfunction. These simulated associations between covariates and EGFR inhibitor exposure are generally similar to observed findings (30-32). Consistent with FDA guidance advising against co-administration with strong CYP3A4 inhibitors and inducers, dosing with rifampicin (600 mg q.d.) for 7 days prior to the administration of EGFR KIs caused a marked reduction in afatinib, erlotinib and gefitinib exposure. Similarly, co-administration of ketoconazole (400 mg q.d.) caused a moderate increase in afatinib exposure, and marked increases in erlotinib and gefitinib exposures.
The potential applicability of the gefitinib compound profile to optimised KI dosing was demonstrated through simulations performed in 1,000 cancer patients using a dosing regimen based on the recommended fixed dosing protocol for this drug (9). These simulations demonstrated that while the mean trough concentration at day 15 was greater than the established target threshold for this drug (259 versus 200 ng/mL), 448 (45%) patients were predicted to experience sub-therapeutic exposure (i.e., trough concentrations <200 ng/mL) at this time (Figure 5), with some patients experiencing as low as 25% of the established target steady-state threshold exposure. These simulated exposure profiles using a well validated full-PBPK gefitinib profile emphasise the need to individualise gefitinib dosing in order to maximise the benefit of this drug.
In conclusion, we have developed and validated robust mechanistic models with the capacity to describe EGFR KI exposure and the impact of covariates on exposure. These models may be applied to inform the impact of different dosing regimens on EGFR KI exposure, the potential impact of poor compliance on EGFR KI efficacy, the need to perform bridging studies when introducing EGFR KIs to new international markets, and the potential impact of DDIs on EFGR KI exposure. In the future, with further enhancement of the Simcyp population profiles, these profiles may also be applied to assist in optimising dosing in individual patients under the Virtual Twin framework.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Precision dosing of targeted anticancer drugs”. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.16). The series “Precision dosing of targeted anticancer drugs” was commissioned by the editorial office without any funding or sponsorship. AR served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Translational Cancer Research. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval was obtained from the Southern Adelaide Clinical Research Ethics Committee for the use of human liver tissue in drug disposition studies in vitro. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol 2008;21:S16-22. [Crossref] [PubMed]
- He M, Capelletti M, Nafa K, et al. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res 2012;18:1790-7. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Yang JJ, Zhang XC, Su J, et al. Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res 2014;20:1383-92. [Crossref] [PubMed]
- Stewart EL, Tan SZ, Liu G, et al. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res 2015;4:67-81. [PubMed]
- Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med 2011;3:90ra59 [Crossref] [PubMed]
- Shah NT, Kris MG, Pao W, et al. Practical management of patients with non-small-cell lung cancer treated with gefitinib. J Clin Oncol 2005;23:165-74. [Crossref] [PubMed]
- Shao J, Markowitz JS, Bei D, et al. Enzyme- and Transporter-Mediated Drug Interactions with Small Molecule Tyrosine Kinase Inhibitors. J Pharm Sci 2014;103:3810-33. [Crossref] [PubMed]
- Rowland A, Van Dyk M, Mangoni A, et al. Kinase inhibitor pharmacokinetics: Comprehensive summary and roadmap for addressing inter-individual variability in exposure Exp Opin Drug Metab Toxicol 2017;13:31-49.
- Scheffler M, Di Gion P, Doroshyenko O, et al. Clinical Pharmacokinetics of Tyrosine Kinase Inhibitors. Clin Pharmacokinet 2011;50:371-403. [Crossref] [PubMed]
- Di Gion P, Kanefendt F, Lindauer A, et al. Clinical Pharmacokinetics of Tyrosine Kinase Inhibitors. Clin Pharmacokinet 2011;50:551-603. [Crossref] [PubMed]
- de Wit D, Guchelaar HJ, den Hartigh J, et al. Individualized dosing of tyrosine kinase inhibitors: are we there yet? Drug Discov Today 2015;20:18-36. [Crossref] [PubMed]
- van Erp NP, Gelderblom H, Guchelaar HJ. Clinical pharmacokinetics of tyrosine kinase inhibitors. Cancer Treat Rev 2009;35:692-706. [Crossref] [PubMed]
- Emoto C, Murayama N, Rostami-Hodjegan A, et al. Methodologies for investigating drug metabolism at the early drug discovery stage: prediction of hepatic drug clearance and P450 contribution. Curr Drug Metab 2010;11:678-85. [Crossref] [PubMed]
- Rowland Yeo K, Jamei M, Yang J, et al. Physiologically based mechanistic modelling to predict complex drug-drug interactions involving simultaneous competitive and time-dependent enzyme inhibition by parent compound and its metabolite in both liver and gut - the effect of diltiazem on the time-course of exposure to triazolam. Eur J Pharm Sci 2010;39:298-309. [Crossref] [PubMed]
- Rostami-Hodjegan A, Tucker GT. Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat Rev Drug Discov 2007;6:140-8. [Crossref] [PubMed]
- Bowalgaha K, Elliot DJ, Mackenzie PI, et al. S-Naproxen and desmethylnaproxen glucuronidation by human liver microsomes and recombinant human UDP-glucuronosyltransferases (UGT): role of UGT2B7 in the elimination of naproxen. Br J Clin Pharmacol 2005;60:423-33. [Crossref] [PubMed]
- Boase S, Miners JO. In vitro-in vivo correlations for drugs eliminated by glucuronidation: Investigations with the model substrate zidovudine. Br J Clin Pharmacol 2002;54:493-503. [Crossref] [PubMed]
- Lowry OH, Rosebrough NJ, Farr AL, et al. Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265-75. [PubMed]
- Rowland A, Elliot DJ, Knights KM, et al. The "albumin effect" and in vitro-in vivo extrapolation: Sequestration of long-chain unsaturated fatty acids enhances phenytoin hydroxylation by human liver microsomal and recombinant cytochrome P450 2C9. Drug Metab Dispos 2008;36:870-7. [Crossref] [PubMed]
- Korprasertthaworn P, Rowland A, Lewis BC, et al. Effects of amino acid substitutions at positions 33 and 37 on UDP-glucuronosyltransferase 1A9 (UGT1A9) activity and substrate selectivity. Biochem Pharmacol 2012;84:1511-21. [Crossref] [PubMed]
- Jones HM, Houston JB. Substrate depletion approach for determining in vitro metabolic clearance: time dependencies in hepatocyte and microsomal incubations. Drug Metab Dispos 2004;32:973-82. [Crossref] [PubMed]
- Jamei M, Marciniak S, Feng K, et al. The Simcyp® Population-based ADME Simulator. Expert Opin Drug Metab Toxicol 2009;5:211-23. [Crossref] [PubMed]
- Dong JQ, Chen B, Gibbs MA, et al. Applications of Computer-Aided Pharmacokinetic and Pharmacodynamic Methods from Drug Discovery Through Registration. Curr Comput Aided Drug Des 2008;4:54-66. [Crossref]
- Howgate EM, Rowland Yeo K, Proctor NJ, et al. Prediction of in vivo drug clearance from in vitro data. I: impact of inter-individual variability. Xenobiotica 2006;36:473-97. [Crossref] [PubMed]
- Polasek TM, Polak S, Doogue MP, et al. Assessment of inter-individual variability in predicted phenytoin clearance. Eur J Clin Pharmacol 2009;65:1203-10. [Crossref] [PubMed]
- Wattanachai N, Polasek TM, Heath TM, et al. In vitro-in vivo extrapolation of CYP2C8-catalyzed paclitaxel 6alpha-hydroxylation: effects of albumin on in vitro kinetic parameters and assessment of interindividual variability in predicted clearance. Eur J Clin Pharmacol 2011;67:815-24. [Crossref] [PubMed]
- Cheeti S, Budha NR, Rajan S, et al. A physiologically based pharmacokinetic (PBPK) approach to evaluate pharmacokinetics in patients with cancer. Biopharm Drug Dispos 2013;34:141-54. [Crossref] [PubMed]
- van Dyk M, Miners J, Kichenadasse G, et al. A novel approach for the simultaneous quantification of 18 small molecule kinase inhibitors in human plasma: a platform for optimised KI dosing. J Chromatogr B Analyt Technol Biomed Life Sci 2016;1033:17-26. [Crossref] [PubMed]
- FDA. Afatinib (GILOTRIF) clinical pharmacology and biopharmaceutics review. Center for Drug Evaluation and Research 2014. Application Number: 201292Orig1s000.
- FDA. Erlotinib (TARCEVA) clinical pharmacology and biopharmaceutics review. Center for Drug Evaluation and Research 2014. Application Number: 021743.
- FDA. Gefitinib (IRESSA) clinical pharmacology and biopharmaceutics review. Center for Drug Evaluation and Research 2014. Application Number: 206995Orig1s000.
- Zhao YY, Li S, Zhang Y, et al. The relationship between drug exposure and clinical outcomes of non-small cell lung cancer patients treated with gefitinib. Med Oncol 2011;28:697-702. [Crossref] [PubMed]
- Gao B, Yeap S, Clements A, et al. Evidence for therapeutic drug monitoring of targeted anticancer therapies. J Clin Oncol 2012;30:4017-25. [Crossref] [PubMed]
- Klümpen HJ, Samer CF, Mathijssen RH, et al. Moving towards dose individualization of tyrosine kinase inhibitors. Cancer Treat Rev 2011;37:251-60. [Crossref] [PubMed]
- Rakhit A, Pantze MP, Fettner S, et al. The effects of CYP3A4 inhibition on erlotinib pharmacokinetics: computer-based simulation (SimCYP) predicts in vivo metabolic inhibition. Eur J Clin Pharmacol 2008;64:31-41. [Crossref] [PubMed]
- Yamazaki S, Johnson TR, Smith BJ. Prediction of Drug-Drug Interactions with Crizotinib as the CYP3A Substrate Using a Physiologically Based Pharmacokinetic Model. Drug Metab Dispos 2015;43:1417-29. [Crossref] [PubMed]
- Deeken JF, Beumer JH, Anders NM, et al. Preclinical assessment of the interactions between the antiretroviral drugs, ritonavir and efavirenz, and the tyrosine kinase inhibitor erlotinib. Cancer Chemother Pharmacol 2015;76:813-9. [Crossref] [PubMed]


