Microarray analysis reveals altered expression of multiple circular RNAs in the pathogenesis of esophageal squamous cell carcinoma
Introduction
Esophageal carcinoma is the eighth most prevalent and sixth most fatal malignancy worldwide (1). In China, esophageal carcinoma is the fourth most common cancer with a much higher incidence rate than in Western countries (2). Esophageal adenocarcinoma (EAC) and squamous cell carcinoma (ESCC) are the two main types of esophageal carcinoma. Unlike Western countries, where EAC is rapidly increasing in incidence and is the dominating type (3), ESCC is the most common type of esophageal cancer in China (2). It is well known that the predilection site and biological behavior differ between ESCC and EAC, indicating that a distinctive molecular biological mechanism is involved in the development and metastasis of ESCC. Many controversies exist with respect to therapy of ESCC, including the extent of lymphadenectomy (4) and the clinical value of preoperative chemoradiotherapy or chemotherapy alone (5). These unclear issues in clinical practice are the manifestation of an incomplete understanding of the molecular and genetic basis of ESCC and may partially be responsible for the poor prognosis of ESCC at present. The development of new biomarkers that could act as diagnostic indicators, therapeutic targets, or prognosis predictors is eagerly anticipated to improve the diagnosis and treatment of ESCC.
Circular RNAs (circRNAs) are an emerging class of endogenous noncoding RNAs characterized by a particular covalently closed-loop molecular structure resulting from a 3’ to 5’ end joining that is stable and resistant to degradation. circRNAs were first discovered in eukaryotic cells in 1979 by Hsu and Coca-Prados using electron microscopy (6); however, research on circRNA in the following decades has been slow due to limited understanding and lack of specific detection technology. In the past few years, the introduction of high-throughput RNA sequencing technologies has made it possible to directly detect circRNA sequences in the whole genome, which has shed new light on circRNA studies. Recent studies revealed that circRNAs are widely involved in a variety of diseases such as neurodegenerative diseases (7), hematological malignancies (8), and gastric cancer (9) through miRNA sponging or other regulatory roles (10,11).
At present, there are only a small number of studies reporting the function of circRNA in ESCC (12-14), and the specific molecular mechanism by which circRNAs regulate ESCC is still unclear. In this study, we performed a circRNA microarray analysis to identify differentially expressed circRNAs between ESCC and adjacent non-neoplastic tissues and then explored their underlying function by prediction of circRNA/miRNA interactions. Our results suggest that multiple circRNAs may be involved in the pathogenesis of ESCC.
Methods
Patients and sample collection
ESCC specimens were obtained from 5 patients (3 males and 2 females with age ranging from 51 to 58 years) with postoperative pathologic stage III disease (IIIa in 4 and IIIc in 1 patient) who underwent minimally invasive esophagectomy and three-field lymph node dissection between May 2016 and June 2016 at Fujian Union Hospital (Table 1). The control group consisted of adjacent non-neoplastic tissues collected from the same patients. All samples were preserved in liquid nitrogen immediately after resection and then transferred to the Institute of Cardiothoracic Surgery of Fujian Union Hospital for storage at −80 °C.
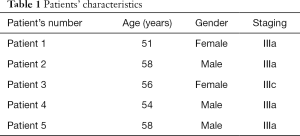
Full table
RNA extraction and sample quality control
Total RNA was extracted from five pairs of tumor and adjacent non-neoplastic tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The purity and concentration of total RNA samples were measured and quantified by NanoDrop ND-1000 (NanoDrop Technologies/Thermo Scientific, Wilmington, DE, USA). The integrity of RNA was tested by electrophoresis on a denaturing agarose gel.
RNA labeling and hybridization
We completed RNA labeling and array hybridization following the manufacturer’s protocol (Arraystar Inc., Rockville, MD, USA). Extracted total RNAs were treated with RNase R (Epicentre, Madison, WI, USA) so that linear RNAs were removed and circular RNAs were enriched. The enriched circular RNAs were then amplified and transcribed into fluorescent cRNA with random primers following the protocol of Arraystar Super RNA Labeling Kit. RNeasy Mini Kit (Qiagen, Hilden, Germany) was used to purify the labeled cRNAs (pmol Cy3/µg cRNA) and NanoDrop ND-1000 was used to determine the concentration and specific activity of the labeled cRNAs in order to assess labeling efficiency. Next, 5 µL of 10× blocking agent and 1 µL of 25× fragmentation buffer were added into 1 µg of each labeled cRNA sample, followed by heating the mixture at 60 °C for 30 min to fragment the labeled cRNA. Finally, the labeled cRNA samples were diluted by adding 25 µL of 2× hybridization buffer. A 50 µL volume of the hybridized solution was then dispensed into a gasket slide, which was assembled with a circRNA expression microarray slide. After incubation at 65 °C for 17 hours in an Agilent hybridization oven, the hybridized arrays were washed and fixed and then scanned by the Agilent Scanner to generate circRNAs expression profiles for microarray data analysis.
Microarray data analysis
Raw data extraction was performed after the scanned images were imported into Agilent Feature Extraction software. Quantile normalization of the raw data and subsequent data processing were performed using the limma package in R software. Low intensity filtering was then performed, and the circRNAs with flags in “Present” or “Marginal” (“All Targets Value”) in at least 5 out of the 10 samples were selected for further analyses. To compare differences in circRNAs expression profile between the ESCC and adjacent non-tumor tissue groups, we calculated the “fold change” (i.e., the ratio of the group averages) between the groups for each circRNA. t-test was used to estimate statistical significance of the differences. Significant differential expression of circRNAs was defined by fold change >1.5 and P value <0.05 in this study. We selected Data/Sort & Filter function in Microsoft Excel to filter the acquired data and rank the differentially expressed circRNAs based on parameters such as fold change and P value.
qRT-PCR validation of candidate circRNAs
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to validate the differentially expressed circRNAs between ESCC tissues and control tissues. The extracted total RNA was reverse transcribed into cDNA using Super Script III Reverse Transcriptase (Invitrogen) following a standard protocol. The relative expression of circRNAs was determined using the ViiA 7 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). circRNA levels were normalized using β-actin as the internal control. The data were calculated by the 2−ΔΔCt method, and quantitative PCR was performed in triplicate. The specific primers used in this study are listed in Table 2.

Full table
Prediction of circRNA/miRNA interaction
An in-house miRNA target prediction software from Arraystar, which was based on TargetScan (15) & miRanda (16) applications and databases, was used to facilitate the prediction of circRNA/microRNA interaction, and the differentially expressed circRNAs between groups were annotated in detail using the circRNA/miRNA interaction information.
Statistical analyses
All data are presented as the mean ± standard deviation (SD). Student’s t-test was used or comparisons between groups. A P value of <0.05 was considered statistically significant.
Results
Differentially expressed circRNAs based on microarray analysis
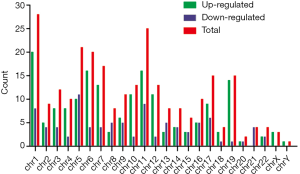
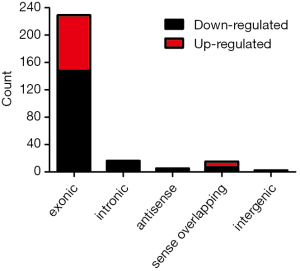
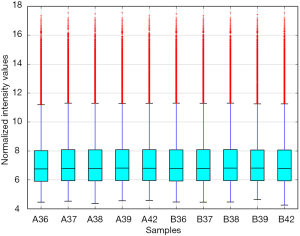
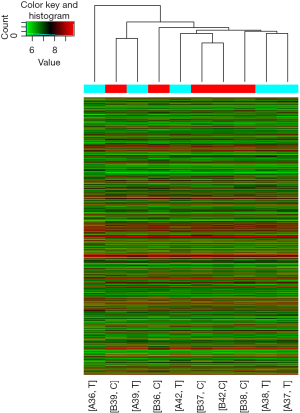
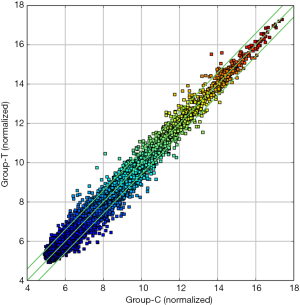
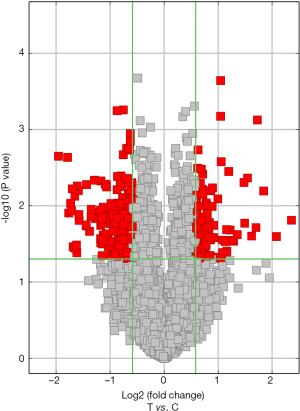
In this study, in total, 12,652 circRNAs were detected by the Arraystar Human circRNA Array. We used a box plot to examine and compare the distributions of expression values for the samples after normalization. As shown in Figure 1, the distribution of circRNAs was similar among all tested samples. Hierarchical clustering was performed to analyze circRNA expression in order to formulate hypotheses about the relationships among samples. The hierarchical clustering showed distinguishable circRNA expression profiles among the samples (Figure 2). A scatter plot was used to facilitate assessment of the variation in circRNA expression between the ESCC and non-neoplastic tissue groups (Figure 3). Volcano plots were then used to determine differential expression between the two groups, with red dots representing differentially expressed circRNAs that reached statistical significance (Figure 4). We identified 267 circRNAs that were differentially expressed in ESCC compared with non-neoplastic tissues, including 92 upregulated and 175 downregulated circRNAs (fold change >1.5, P value <0.05). The differentially expressed circRNAs are categorized and summarized in Figures 5 and 6.
Validation of selected circRNAs using qRT-PCR
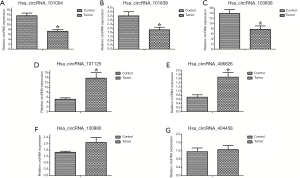
We randomly selected seven significantly differentially expressed circRNAs for validation in all samples by qRT-PCR, and among them, five circRNAs exhibited expression patterns consistent with the microarray data. These five circRNAs included two upregulated (hsa_circRNA_101125 and hsa_circRNA_406826) and three downregulated (hsa_circRNA_101004, hsa_circRNA_103836 and hsa_circRNA_101839) circRNAs (Table 3). The results of qRT-PCR validation are shown in Figure 7.

Full table
Prediction of circRNA/microRNA interactions
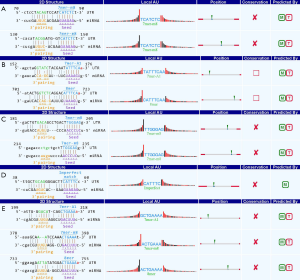
According to the ceRNA hypothesis and a previous report (10), circRNAs may interact with miRNAs through miRNA response elements (MREs) and thus function as miRNA sponges. Therefore, we used an in-house miRNA target prediction software from Arraystar to predict miRNAs that may bind to the selected circRNAs. The information on circRNA/miRNA interactions for the five confirmed circRNAs including the most likely potential targeted miRNAs is summarized in Figure 8.
Discussion
In the classic gene expression model, known as the central dogma, the expression and adjustment of genetic and functional information within the genome are mediated by linear RNAs. The emergence of circRNAs represented both a challenge and a supplement to the central dogma. Recent studies have shown that circRNAs are highly abundant in the cytoplasm of eukaryotic cells, at levels comparable to or even exceeding those of their canonical linear isoforms (17,18). Moreover, circRNAs are structurally stable, highly evolutionarily conserved, and tissue specific, with multiple regulatory functions (18-22). The mechanism by which circRNAs exert regulatory functions is far from clear. Exonic circRNAs, the most abundant type, have been found to function as miRNA sponges. ciRS-7, one of the best known circRNAs, contains more than 70 MREs. ciRS-7 can specifically bind to miR-7 and strongly suppress miR-7 activity (10,23). Likewise, the sex-determining region Y (SRY) serves as a miR-138 sponge (10,24). In the past few years, hsa_circ_001569 (25), circPVT1 (26), circTCF25 (27) and cir-ITCH (12) were discovered as new circRNAs with miRNA sponge function. These circRNAs have been shown to be involved in the development of several kinds of carcinoma by sequestering tumor-related miRNAs. In addition, exonic circRNAs can also function by regulating transcription (28) and interacting with RNA binding proteins (29), but elucidation of specific mechanisms requires further investigation. For other circRNA types, such as intronic circRNAs and exon-intron circRNAs (EIciRNAs), there have only been a few relevant studies. Existing reports reveal that these circRNAs regulate transcription by enhancing the expression of their parental gene (11,30). The characteristics mentioned above qualify circRNAs as a promising target for the diagnosis and treatment of diseases.
There are very few studies on circRNAs in ESCC. Li et al. (12) discovered that cir-ITCH was expressed at low levels in ESCC compared with peritumoral tissues. Furthermore, it was shown to function as a sponge for miR-7, miR-17, and miR-214, subsequently increasing the level of ITCH and thus playing an inhibitory role in ESCC by regulating the Wnt pathway. Su et al. (13) performed circRNA microarray and bioinformatics analyses to compare expression profiles of circRNA in radioresistant and non-radioresistant ESCC cells. They reported a comprehensive expression and functional profile of differentially expressed circRNAs in radioresistant esophageal cancer cells, among which circRNA_001059 and circRNA_000167 were identified as the two largest nodes in the circRNA/microRNA co-expression network. Xia et al. (14) found that hsa_circ_0067934 was upregulated in ESCC tumor tissues, and showed that in vitro silencing of hsa_circ_0067934 resulted in the inhibition of proliferation and migration of ESCC cells and cell cycle arrest. This suggested that circ_0067934 has potential as a novel biomarker and therapeutic target of ESCC. Although the specific regulatory mechanism of circRNA in ESCC was unclear, the inspiring results of these studies confirmed that circRNAs play a role in the process of ESCC pathogenesis.
In this study, we compared circRNA expression profiles between ESCC and adjacent non-neoplastic tissues using Arraystar Human Circular RNA Microarrays to investigate the mechanism of ESCC pathogenesis. Multiple differentially expressed circRNAs were observed and validated in the ESCC tissues compared with the adjacent non-neoplastic tissues. Our results revealed that these dysregulated circRNAs might be involved in the process of ESCC pathogenesis, especially in the case of hsa_circRNA_101125 and hsa_circRNA_406826, which showed significantly upregulated expression in the ESCC tissues. According to the prediction results for circRNA/microRNA interactions, hsa_circRNA_101125 has a potential binding site for miR-143-3p and thus might function as a miR-143-3p sponge. miR-203a-3p was a predicted target of hsa_circRNA_406826, which might sequester miR-203a-3p through miRNA response elements. According to previous reports, both miR-143-3p and miR-203a-3p are downregulated in ESCC tissues (31,32), and further in vivo and in vitro studies have provided strong evidence that both miRNAs act as tumor suppressors in ESCC. Enforced expression of miR-143-3p and miR-203a-3p inhibited growth, proliferation, migration and invasion of ESCC cells through different mechanisms, while their reduced expression was associated with poor patient survival (33-43). Specific binding of hsa_circRNA_101125 and miR-143-3p or that of hsa_circRNA_406826 and miRNA-203a-3p might relieve the suppression of tumor-related genes by miRNAs, thus promoting the occurrence and development of ESCC. Regarding the three circRNAs with downregulated expression, we believe that miRNA sponging may not be their main function in ESCC pathogenesis, and the mechanism associated with their low expression level remains to be clarified.
Conclusions
In summary, we detected differential circRNA expression profiles between ESCC and adjacent non-neoplastic tissues. Our results demonstrated that multiple circRNAs were aberrantly expressed in ESCC tissues and might be involved in the process of ESCC pathogenesis. In particular, hsa_circRNA_101125 and hsa_circRNA_406826 were significantly upregulated in ESCC, and these circRNAs might serve as potential diagnostic or therapeutic markers in ESCC. Further studies are needed to clarify their specific regulatory mechanisms.
Acknowledgments
Funding: This work was supported by the Science and Technology Key Project of Fujian Province, China (Grant Number: 2014Y0024) and Youth Fund of Fujian Health Department (Grant Number: 2012-1-18).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.39). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Fujian Medical University Union Hospital (No. 2016ky012), and written informed consent was obtained from all patients. The experiments were performed according to the Ethical Guidelines for Human Genome/Gene Research issued by the Chinese Government.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol 2009;24:729-35. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA-Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ma GW, Situ DR, Ma QL, et al. Three-field vs two-field lymph node dissection for esophageal cancer:a meta-analysis. World J Gastroenterol 2014;20:18022-30. [Crossref] [PubMed]
- Kumagai K, Rouvelas I, Tsai JA, et al. Meta-analysis of postoperative morbidity and perioperative mortality in patients receiving neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal and gastro-oesophageal junctional cancers. Brit J Surg 2014;101:321-38. [Crossref] [PubMed]
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. [Crossref] [PubMed]
- Kumar L. Circular RNAs: the Emerging Class of Non-coding RNAs and Their Potential Role in Human Neurodegenerative Diseases. Mol Neurobiol 2016; [Epub ahead of print]. [PubMed]
- Bonizzato A, Gaffo E, Te Kronnie G, et al. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J 2016;6:e483 [Crossref] [PubMed]
- Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015;444:132-6. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015;6:6001-13. [Crossref] [PubMed]
- Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016;14:225. [Crossref] [PubMed]
- Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep 2016;6:35576. [Crossref] [PubMed]
- Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol 2003;5:R1. [Crossref] [PubMed]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012;13:271-82. [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. Plos One 2012;7:e30733 [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 2013;19:141-57. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011;30:4414-22. [Crossref] [PubMed]
- Granados-Riveron JT, Aquino-Jarquin G. Does the linear Sry transcript function as a ceRNA for miR-138? The sense of antisense. F1000Res 2014;3:90. [PubMed]
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. [Crossref] [PubMed]
- Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 2017;388:208-19. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Wu BL, Xu LY, Du ZP, et al. MiRNA profile in esophageal squamous cell carcinoma:downregulation of miR-143 and miR-145. World J Gastroenterol 2011;17:79-88. [Crossref] [PubMed]
- Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg 2008;135:255-60. [Crossref] [PubMed]
- Ni Y, Meng L, Wang L, et al. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene 2013;517:197-204. [Crossref] [PubMed]
- Mao Y, Liu J, Zhang D, et al. MiR-143 inhibits tumor progression by targeting FAM83F in esophageal squamous cell carcinoma. Tumour Biol 2016;37:9009-22. [Crossref] [PubMed]
- Liu J, Mao Y, Zhang D, et al. MiR-143 inhibits tumor cell proliferation and invasion by targeting STAT3 in esophageal squamous cell carcinoma. Cancer Lett 2016;373:97-108. [Crossref] [PubMed]
- Takeshita N, Mori M, Kano M, et al. MiR-203 inhibits the migration and invasion of esophageal squamous cell carcinoma by regulating LASP1. Int J Oncol 2012;41:1653-61. [Crossref] [PubMed]
- Yu X, Jiang X, Li H, et al. MiR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev 2014;23:576-85. [Crossref] [PubMed]
- Li J, Shan F, Xiong G, et al. EGF-induced C/EBPβ participates in EMT by decreasing the expression of miR-203 in esophageal squamous cell carcinoma cells. J Cell Sci 2014;127:3735-44. [Crossref] [PubMed]
- Zhang F, Yang Z, Cao M, et al. MiR-203 suppresses tumor growth and invasion and down-regulates MiR-21 expression through repressing Ran in esophageal cancer. Cancer Lett 2014;342:121-9. [Crossref] [PubMed]
- Hu Y, Correa AM, Hoque A, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer 2011;128:132-43. [Crossref] [PubMed]
- Okumura T, Shimada Y, Moriyama M, et al. MicroRNA-203 inhibits the progression of esophageal squamous cell carcinoma with restored epithelial tissue architecture in vivo. Int J Oncol 2014;44:1923-32. [Crossref] [PubMed]
- Zhang K, Dai L, Zhang B, et al. MiR-203 is a direct transcriptional target of E2F1 and causes G1 arrest in esophageal cancer cells. J Cell Physiol 2015;230:903-10. [Crossref] [PubMed]
- Liu Y, Dong Z, Liang J, et al. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour Biol 2016;37:5621-32. [Crossref] [PubMed]