
Alteration of non-coding RNAs expression pattern in metastasis process of esophageal squamous cell carcinoma
Introduction
Esophageal cancer (EC) is the common malignancy in digestive tract and ranks the sixth leading cause in male and ninth leading cause of cancer-related mortality in female in 2012 worldwide (1). Patients with EC are frequently diagnosed at advanced stages, with 5 years survival rate less than 10%, deprived of the chance of surgical resection for long-term survival (2,3).
Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) are the two histological subtypes of EC. Smoking, alcohol drinking and low intake of fruits and vegetables increase the occurrence risk of ESCC (4).
In currently, the etiological factors of ESCC are unclearly. It is reported that aberrant expression of non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) regulate multiple tumorigenic processes, including proliferation, invasion, metastasis and prognosis in ESCC. Decreased miR-630 induces ECCC cell proliferation, invasion, metastasis, EMT and poor overall survival of patients with ESCC (5). High plasma concentration of oncogenic miR-21 is an independent risk factor of chemo-resistance in ESCC (6). miR-143-3p is decreased in ESCC tissues and ectopic expression of miR-143-3p inhibits ESCC cell proliferation and induces cell apoptosis (7). Except of miRNAs, lncRNAs also play vital roles in ESCC tumorigenesis. Up-regulation of lncRNA BANCR, PCAT-1 and BC200 is correlated with advanced TNM stage, lymph node metastasis and shorter survival (8-10). LOC100130476 is significantly down-regulated in ESCC cell lines and tissues; up-regulation of LOC100130476 inhibits ESCC cell proliferation and invasiveness (11). lncRNA POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with a significantly decreased risk for ESCC (12).
At present, the tumorigenesis and metastasis mechanism of ESCC is largely unknown. In this study, dysregulated lncRNAs, miRNAs and mRNAs were identified in ESCC metastasis. Our study investigated non-coding RNA expression pattern and might provide valuable information for exploring pathogenesis and metastasis mechanism in ESCC.
Methods
Patients and samples
Seven ESCC patients with metastasis and seven ESCC patients without metastasis in the Daping Hospital were enrolled in our study. Among which, three ESCC patients with metastasis and three ESCC patients without metastasis were enrolled for RNA-sequencing; four ESCC patients with metastasis and four ESCC patients without metastasis were enrolled for quantitative real-time polymerase chain reaction (qRT-PCR). Primary tumor tissues of ESCC patients with metastasis and non-metastasis were obtained from esophagectomy. All these patients were diagnosed as ESCC based on postoperative pathology and none of them, received chemo- or radiotherapy before esophagectomy. The detailed information of patients was displayed in Tables 1 and 2.
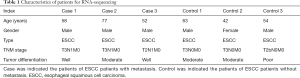
Full table
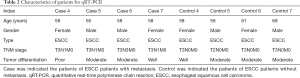
Full table
Ethics
This work was approved by the Ethics Committee of the Daping Hospital (2015-035) and informed written consent was obtained from all patients. The research complied with the principles of the Declaration of Helsinki.
Library preparation and high-throughput sequencing
Three primary tumor loci of ESCC patients without metastasis (primary group) and three primary tumor loci of ESCC patients with metastasis (metastasis group) were pooled for RNA-sequencing, respectively. Total RNA of collected specimens was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Then cDNA libraries were constructed according to the manufacture instruction. Likewise, cDNA libraries of small RNA were constructed. Lastly, each library was loaded into one lane of the illumine hiseq 4,000 for sequencing.
Data preprocessing
The raw image data obtained from high-throughput RNA-sequencing was translated into raw FASTQ sequence data by Base Calling. Nucleotides with a quality score <20 were trimmed from the end of the sequence and N base rate of raw reads more than 10% were discarded using Cutadapt 1.9.1. Finally, clean reads were obtained (13). TopHat was used to align the clean reads with the human reference genome, Ensemble GRCh38 vs. 84 (hg19) by using (14). Fragments per kilobase of exon per million fragments mapped (FPKM) was used to decipher the transcription abundance of long non-coding RNA (lncRNA) and protein coding mRNA (mRNA), which was quantified by cuffquant and cuffnorm. miRDeep2 was used to quantified the transcription abundance of miRNAs (15).
Differentially expressed genes analysis
The differentially expressed lncRNAs (DELs) and differentially expressed mRNA (DEMs) were identified in metastasis group compared with primary group via Cuffdiff. lncRNAs and mRNAs with FDR <0.05 and |log2FC >1| was selected as DELs and DEMs. In addition, differentially expressed miRNAs (DEMIs) in metastasis group compared with primary group were identified using DEGseq package in R language, with the threshold of FDR <0.001 and |log2FC >1|.
Identification of target genes of DEMIs
miRWalk database (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), was used to predict the target genes of DEMIs (16). In our study, 6 algorithms including RNA22, miRanda, miRDB, miRWalk, PICTAR and Targetscan in miRWalk was used. Moreover, the genes, predicted by more than 4 out of 6 algorithms, were as considered as the target gene of the DEMIs. DEMIs-target gene interacting pairs in a negative manner were subjected to construct DEMI-target gene interaction network, visualized by Cytoscape (17).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
In order to obtain insight into the biological functions and signaling pathways of dysregulated mRNAs in metastasis group involved in, GeneCoDis3 (http://genecodis.cnb.csic.es/analysis) analysis (18), including GO function and KEGG pathway enrichment were conducted. Items with FDR <0.05 was filed as significant enrichments.
qRT-PCR
Total RNA of metastasis group and primary group tissues were extracted by using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacture instructions.
FastQuant cDNA and miRcute Plus (Tiangen, Beijing, China) and miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China) was used to synthesize the cDNA of mRNA and miRNA, respectively. qRT-PCR reactions were performed by using SuperReal PreMix Plus SYBR Green Kit (Tiangen, Beijing, China) and miRcute Plus miRNA qPCR Detection Kit (Tiangen, Beijing, China) on Applied Biosystems 7500 (Applied Biosystems, Foster City, CA, USA). GAPDH and U6 were used as internal control for mRNA and miRNA detection, respectively. The relative expression of candidate genes was calculated by using the 2-ΔΔCT equation methods (19). The PCR primers used in our study were shown in Table S1. At least triple experiments were subjected to qRT-PCR verification.
Validation the expression of selected DELs, DEMIs and DEMs in the Cancer Genome Atlas (TCGA) database
As a public funded project, TCGA (https://tcga-data.nci.nih.gov/tcga/) stores multidimensional data of various human tumors at the DNA, RNA and protein levels. A TCGA illumine hiseq data consisted of 32 metastasis ESCC tissues and 40 primary ESCC tissues were used to validate the expression of three DELs (LINC00668, XIST and SNHG5) and nine DEMs (AKR1B10, SOX5, CYP4F11, SLC7A11, HLA-C, ICAM2, CLDN8, CTLA4 and KRT19) between metastasis and primary ESCC tissues. A TCGA illumine hiseq data consisted of 38 metastasis ESCC tissues and 48 primary ESCC tissues were used to validate the expression of selected three DEMIs (miR-224, miR-229 and miR-200a) between metastasis and primary ESCC tissues.
Statistical analysis
Mean ± standard deviation and independent-samples t-test was used in the statistical analysis. P<0.05 was considered as significant difference. * indicated P<0.05; ** indicated P<0.01 and *** indicated P<0.001.
Results
Identification of DELs and DMEs in metastasis group tissues compared with primary group tissues
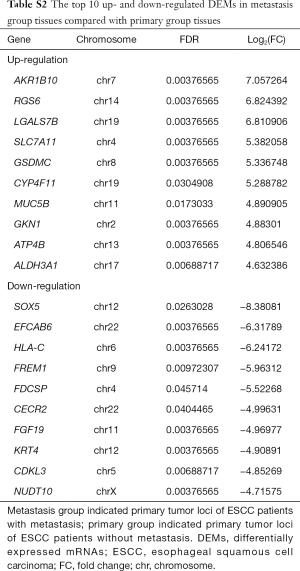
Total of 43 DELs (9 up- and 34 down-regulated) and 205 DEMs (49 up- and 156 down-regulated) were identified in metastasis group tissues based on the threshold of FDR <0.05 and abs (log2FC)≥1. LOC105378306 and LINC01206 were the most obviously up- and down-regulated DEL in metastasis group tissues, respectively (Table 3). In addition, AKR1B10 and SOX5 were the most obviously up- and down-regulated DEM in metastasis group tissues, respectively (Table S2).
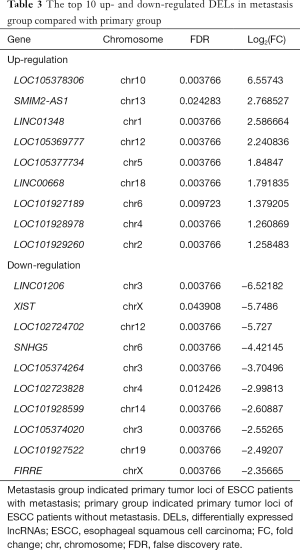
Full table
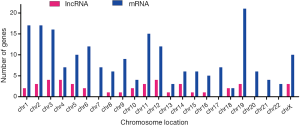
In our study, 43 identified DELs were distributed in chromosomes X and 17 autosomes, apart from chromosome 7, 17, 20, 21 and 22; 205 DEMs were distributed in all autosomes and chromosomes X (Figure 1).
Identification of DEMIs in metastasis group tissues
Total of 128 DEMIs (9 up- and 119 down-regulated) were identified in metastasis group tissues compared with primary group tissues based on the threshold of FDR <0.001 and abs (log2FC)1.5. The hsa-miR-508-3p, hsa-miR-224-5p and hsa-miR-514a-3p were the three significantly up-regulated DEMIs in metastasis group tissues; hsa-miR-3664-3p, hsa-miR-449b-5p and hsa-miR-548ah-3p were the three highly significantly down-regulated DEMIs in ESCC (Table 4).

Full table
DEMIs-DMEs interaction network
The target genes of 128 identified DEMIs, predicted through miRWalk database, were overlapped with 205 DEMs in metastasis group tissues and DEMIs-DMEs interaction pairs were distinguished, which were visualized using Cytoscape. As shown in Figure 2, DEMIs-DEMs interaction network consisted of 145 nodes and 214 edges, which involved in 88 DEMIs and 57 DEMs. The hsa-miR-495-3p, hsa-miR-200b-5p and hsa-miR-200a-5p had the highest connectivity with DEMs, regulated 8, 6 and 6 DEMs, respectively. SLC7A11, GDNF and SRXN1 were regulated by 37, 15 and 13 DEMIs, respectively.
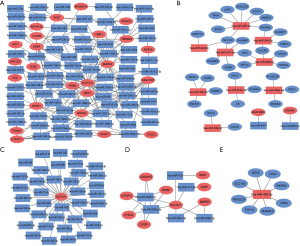
GO and KEGG pathway enrichment
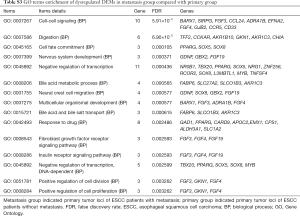
To predict the functions of DEMs targeted by DEMIs, GO and KEGG pathway were conducted to demonstrate it. 205 DEMs were significantly enriched in 14 signaling pathways (Table 5), such as cytokine-cytokine receptor interaction (KEGG: 04060), MAPK signaling pathway (KEGG: 04010), pathways in cancer (KEGG: 05200), chemokine signaling pathway (KEGG: 04062) and cell adhesion molecules (CAMs, KEGG: 04514). In addition, 205 DEMs were significantly enriched in cell-cell signaling, digestion, cell fate commitment, positive regulation of cell division and positive regulation of cell proliferation of biological process (Table S3).

Full table
qRT-PCR validation of the expression level of representative genes
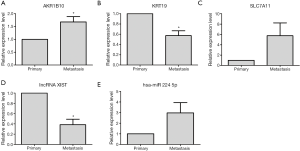
In order to validate the expression level of dysregulated genes in metastasis group tissues based on bioinformatics analysis, qRT-PCR was applied. Five dysregulated genes, including 1 lncRNA XIST, 3 DEMs (AKR1B10, KRT19, SLC7A11) and 1 DEMIs hsa-miR-224-5p, were chose for qRT-PCR verification in 4 metastasis group tissues and 4 primary group tissues. As Figure 3A,B shown, AKR1B10 and KRT19 were significantly up- and down-regulated in metastasis group compared with primary group; SLC7A11 had the up-regulated tendency in metastasis group (Figure 3C). In Figure 3D,E, lncRNA XIST was significantly down-regulated and DEMIs hsa-miR-224-5p had the up-regulated tendency in metastasis group compared with primary group.
Validation the expression of selected DELs, DEMIs and DEMs in TCGA database
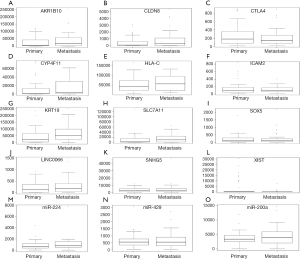
The relative expression of selected DEMs, DEMIs and DELs between metastasis and primary ESCC tissues in the TCGA illumine hiseq data were shown in Figure 4. Four DEMs (AKR1B10, CYP4F11, SLC7A11 and CLDN8), lncRNA LINC00668 and miR-224 were up-regulated while five DEGs (SOX5, HLA-C, ICAM2, CTLA4 and KRT19), two DELs (XIST and SNHG5) and two DEMIs (miR-429 and miR-200a) were down-regulated in metastasis ESCC tissues compared to primary metastasis. Except for HLA-C, SNHG5, miR-429 and miR-200a, the expression of other 11 one was consistent with our RNA-sequencing results, generally.
Discussion
Dysregulated DELs, DEMIs and DEMs were identified in ESCC metastasis group compared with primary group. In order to validate our bioinformatics analysis, the expression of selected DEMs, DEMIs and DELs were validated by qRT-PCR and TCGA illumine hiseq data. The validated expression results of qRT-PCR and TCGA illumine hiseq data were generally in accordance with our RNA-sequencing results which suggested that our RNA-sequencing results were convincing.
The miR-200 family of miRNAs (miR-141, 200a, 200b, 200c and 429) are key regulators/inhibitors of epithelial to mesenchymal transition, which is involved in cancer cell behaviors including cell proliferation, cell cycle and apoptosis (20-22). In our study, all of miR-200 family number including miR-200a-5p, miR-200b-3p, miR-200b-5p, miR-200c-3p, miR-141-3p and miR-429 were significantly down-regulated in metastasis group compared with primary group.
Whereby, miR-200b-5p and miR-200a-5p were the hubs in DEMI-DEM interaction network. It is reported that the expression level of miR-200a, miR-200b, miR-200c, miR-141 and miR-429 are significantly lower in esophageal adenocarcinoma compared with Barrett’s esophagus epithelium (23). The miR-200b suppresses cell invasiveness, cell growth and induces cell cycle arrest in ESCC; whereas, loss of miR-200b promotes cell invasiveness through activating Kindlin-2/integrin β1/AKT pathway in ESCC (24-26). Higher expression of miR-200c in serum patients with ESCC is significantly associated with TNM stage and worse response to platinum-based chemotherapy (27). Increased miR-429 suppressed cell invasiveness and induces cell apoptosis by targeting Bcl-2 and SP-1 in esophageal carcinoma (28). A published article demonstrates miR-141 is significantly over-expressed in ESCC compared with normal tissues, which is disharmony with our in silicon analysis, and over-expression miR-141 contribute to an acquired chemo-resistance in EC-cells (29). Currently, the functions of miR-141 in ESCC were unknown. The expression status of it need to be validated d in ESCC tissues by qRT-PCR in a large sample size of patients with ESCC, in addition, the roles of miR-141 in cell behaviors of ESCC need to be explored in the future work. SLC7A11, with obviously significant up-regulation in metastasis group (Table S2), was targeted by 37 DEMIs, such as 6 abovementioned members of miR-200 family. Our bioinformatics analysis and qRT-PCR verification indicated SLC7A11 was up-regulated in metastasis group compared with primary group. Increased SLC7A11 induces cell growth and is positively correlated with tumor invasiveness and shorter overall survival in patients with glioblastomas (30,31). Along with SCL7A11, CYP4F11, belonged to top 10 up-regulated DEMs in metastasis group (Table S2), was targeted by miR-200b-5p, respectively. CYP4F11 is highly expressed in breast cancer than in adjacent normal tissues, which is associated with Ki67 protein expression (32). However, the biological function of CYP4F11 in ESCC has not been demonstrated. Based on aforementioned information, miR-200 family might play essential roles in tumorigenesis and metastasis process of ESCC by regulating the expression of their target DEMs.
LncRNA LINC00668, XIST and SNHG5 were significantly aberrant expression in metastasis group compared with primary group in ESCC. LINC00668 was ectopically expressed in metastasis group (Table 3). In vivo and in vitro experiments decipher that over-expression of LINC00668, induced by E2F1 transcription factor, enhance cell proliferation and predicts a poor prognosis of patients with gastric cancer (33). The expression level of XIST and SNHG5 was significantly decreased in metastasis group (Table 3). SNHG5 over-expression inhibits cell growth, migration, invasion and metastasis of gastric cancer in vivo and in vitro experiments (34). Moreover, the serum level of SNHG5 is significantly higher in the patients with melanoma than in the normal subjected, which might be a new tumor marker of malignant melanoma (35). XIST is found to be up-regulated and acts as oncogene in a number of cancer types, including non-small cell lung cancer (NSCLC), gastric cancer and glioblastoma (36-38). In NSCLC, increased XIST predicts shorter overall survival and poor prognosis of patients; knockdown of XIST impedes cell proliferation, migration and invasion (36). In gastric cancer, over-expression of XIST is markedly associated with lymph node invasion, distant metastasis and TNM stage in patients; silencing XIST inhibits cell growth and cell metastasis (37). However, the functions of LINC00668, XIST and SNHG5 have not been elucidated in ESCC, which might exert essential functions in tumorigenesis and metastasis of ESCC.
AKR1B10 and SOX5 was the significantly up- and down-regulated DEMs in metastasis group (Table S2). A previous study reports that AKR1B10 is dramatically up-regulated during chemo-resistant induction in gastric cancer cell, which facilitates cell migration and invasiveness, through down-regulating PPARγ and reducing the sensitivity to the chemotherapy (39,40). Protein AKR1B10, a member of aldo keto reductases family, is involved in digestion and is highly expressed in the gastrointestinal tract. AKR1B10 was significantly enriched in digestion of GO terms. SOX3, SOX5, SOX8, the member of SRY-related HGM-box family of transcription factors, were all down-regulated in metastasis group. SOX5 promotes epithelial-mesenchymal transition and cell invasion through regulation of Twist1 in hepatocellular carcinoma (41). In addition, SOX5 and SOX8 were significantly enriched in cell fate commitment in our work. In our study, HLA-C was significantly down-regulated in metastasis group. It is reported that HLA-C is down-regulated at both the protein and mRNA levels in human ESCC, which might attributes to DNA hypermethylation of the promoter region of HLA-C (42). In currently, the mechanisms of AKR1B10, SOX5 and HLA-C in ESCC tumorigenesis and metastasis are ambiguous.
Dysregulated DEMs in metastasis group were highly enriched in 13 KEGG signaling pathways including MAPK signaling pathway, pathways in cancer and CAMs; and significantly enriched in digestion, cell fate commitment, positive regulation of cell division and positive regulation of cell proliferation of biological process. In our work, along with HLA-C, identified DEMs of ICAM2, CLDN8 and CTLA4 were enriched in CAMs pathway, which is involved in a series of biologic processes, including cellular adhesion, cell fate and cell metastasis in cancer (43,44). CAMs are composed of 4 main groups, such as integrin family, the immunoglobulin super family, selectins, and cadherins. In esophageal carcinoma, higher expression of CTLA-4 in the tumor environment is associated with shorter overall survival, which predicts poor prognosis (45). In colon cancer, CAMs pathway associated genes (ICAM2) is modulated by down-regulation WISP1, and protein β-catenin binds with WISP1, exerts the functions of promoting cell proliferation and cell invasiveness (46). CLDN8, an integral membrane protein, forms tight junctions together with occludin. CLDN8 was downregulated in metastasis group in our analysis and it is found to be down-regulated in colorectal tumors compared to adjacent non-tumor tissues (47,48).
Conclusions
In conclusion, we identified the expression profiling of abnormally expressed lnRNAs, miRNAs and mRNAs in ESCC metastasis. Our study indicated that those dysregulated genes including XIST, SNHG5, miR-200 family, AKR1B10 and SOX5 might synergistically contributes to tumorigenesis and metastasis process in ESCC via complex interactions between each other through MAPK signaling pathway, pathways in cancer and CAMs. Our study might lay the foundation for illumination of tumorigenesis and metastasis mechanisms and discovery of potential therapeutic targets in ESCC metastasis. However, pooled RNA-sequencing in our study is a limitation. Although pooled RNA-sequencing is reliable and cost-effective approach to obtain genome-wide information (49), analysis of variation in expression level between individuals was lacked. Sequencing using RNAs from individual ESCC tissues and compare the expression profiling between primary and metastasis ESCC tissues with larger sample size were need.

Full table
Full table
Full table
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.35). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This work was approved by the Ethics Committee of the Daping Hospital (2015-035) and informed written consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA 2012;62:10-29. [PubMed]
- Layke JC, Lopez PP. Esophageal cancer: a review and update. Am Fam Physician 2006;73:2187-94. [PubMed]
- Deschamps C, Nichols FC, Cassivi SD, et al. Long-term function and quality of life after esophageal resection for cancer and Barrett's. The Surgical clinics of North America 2005;85:649-56. [Crossref] [PubMed]
- Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 2015;21:7933-43. [Crossref] [PubMed]
- Jin L, Yi J, Gao Y, et al. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin 2016;48:810-9. [Crossref] [PubMed]
- Komatsu S, Ichikawa D, Kawaguchi T, et al. Circulating miR-21 as an independent predictive biomarker for chemoresistance in esophageal squamous cell carcinoma. Am J Cancer Res 2016;6:1511-23. [PubMed]
- He Z, Yi J, Liu X, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol cancer 2016;15:51. [Crossref] [PubMed]
- Liu Z, Yang T, Xu Z, et al. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother 2016;82:406-12. [Crossref] [PubMed]
- Zhao RH, Zhu CH, Li XK, et al. BC200 LncRNA a potential predictive marker of poor prognosis in esophageal squamous cell carcinoma patients. Onco Targets Ther 2016;9:2221-6. [PubMed]
- Shi WH, Wu QQ, Li SQ, et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol 2015;36:2501-7. [Crossref] [PubMed]
- Guo W, Dong Z, Shi Y, et al. Aberrant methylation-mediated downregulation of long noncoding RNA LOC100130476 correlates with malignant progression of esophageal squamous cell carcinoma. Dig Liver Dis 2016;48:961-9. [Crossref] [PubMed]
- Kang M, Sang Y, Gu H, et al. Long noncoding RNAs POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with decreased risk of esophageal cancer. Tumour Biol 2015;36:6401-8. [Crossref] [PubMed]
- Ghosh S, Chan CK. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol Biol 2016;1374:339-61. [Crossref] [PubMed]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-11. [Crossref] [PubMed]
- Friedländer MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012;40:37-52. [Crossref] [PubMed]
- Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Carmona-Saez P, Chagoyen M, Tirado F, et al. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 2007;8:R3. [Crossref] [PubMed]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat protocols 2008;3:1101-8.
- Smith CM, Watson DI, Michael MZ, et al. MicroRNAs, development of Barrett's esophagus, and progression to esophageal adenocarcinoma. World J Gastroenterol 2010;16:531-7. [Crossref] [PubMed]
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593-601. [Crossref] [PubMed]
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol 2008;5:115-9. [Crossref] [PubMed]
- Smith CM, Watson DI, Leong MP, et al. miR-200 family expression is downregulated upon neoplastic progression of Barrett's esophagus. World J Gastroenterol 2011;17:1036-44. [PubMed]
- Zhang HF, Alshareef A, Wu C, et al. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis 2016;37:858-69. [Crossref] [PubMed]
- Zhang HF, Zhang K, Liao LD, et al. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting Kindlin-2. Carcinogenesis 2014;35:292-301. [Crossref] [PubMed]
- Zhang HF, Alshareef A, Wu C, et al. Loss of miR-200b promotes invasion via activating the Kindlin-2/integrin beta1/AKT pathway in esophageal squamous cell carcinoma: An E-cadherin-independent mechanism. Oncotarget 2015;6:28949-60. [Crossref] [PubMed]
- Yu H, Duan B, Jiang L, et al. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am J Transl Res 2013;6:71-7. [PubMed]
- Wang Y, Li M, Zang W, et al. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) 2013;36:385-94. [Crossref] [PubMed]
- Jin YY, Chen QJ, Xu K, et al. Involvement of microRNA-141-3p in 5-fluorouracil and oxaliplatin chemo-resistance in esophageal cancer cells via regulation of PTEN. Mol Cell Biochem 2016;422:161-70. [Crossref] [PubMed]
- Takeuchi S, Wada K, Toyooka T, et al. Increased xCT expression correlates with tumor invasion and outcome in patients with glioblastomas. Neurosurgery 2013;72:33-41. [Crossref] [PubMed]
- Robert SM, Buckingham SC, Campbell SL, et al. SLC7A11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci Transl Med 2015;7:289ra86 [Crossref] [PubMed]
- Bandala C, Floriano-Sanchez E, Cardenas-Rodriguez N, et al. RNA expression of cytochrome P450 in Mexican women with breast cancer. Asian Pac J Cancer Prev 2012;13:2647-53. [Crossref] [PubMed]
- Zhang E, Yin D, Han L, et al. E2F1-induced upregulation of long noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIs. Oncotarget 2016;7:23212-26. [Crossref] [PubMed]
- Zhao L, Guo H, Zhou B, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene 2016;35:5770-80. [Crossref] [PubMed]
- Ichigozaki Y, Fukushima S, Jinnin M, et al. Serum long non-coding RNA, snoRNA host gene 5 level as a new tumor marker of malignant melanoma. Exp Dermatol 2016;25:67-9. [Crossref] [PubMed]
- Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun 2016;478:811-7. [Crossref] [PubMed]
- Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res 2016;35:142. [Crossref] [PubMed]
- Yao Y, Ma J, Xue Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett 2015;359:75-86. [Crossref] [PubMed]
- Morikawa Y, Kezuka C, Endo S, et al. Acquisition of doxorubicin resistance facilitates migrating and invasive potentials of gastric cancer MKN45 cells through up-regulating aldo-keto reductase 1B10. Chem Biol Interact 2015;230:30-9. [Crossref] [PubMed]
- Matsunaga T, Suzuki A, Kezuka C, et al. Aldo-keto reductase 1B10 promotes development of cisplatin resistance in gastrointestinal cancer cells through down-regulating peroxisome proliferator-activated receptor-gamma-dependent mechanism. Chem Biol Interact 2016;256:142-53. [Crossref] [PubMed]
- Wang D, Han S, Wang X, et al. SOX5 promotes epithelial-mesenchymal transition and cell invasion via regulation of Twist1 in hepatocellular carcinoma. Med Oncol 2015;32:461. [PubMed]
- Nie Y, Yang G, Song Y, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis 2001;22:1615-23. [Crossref] [PubMed]
- Xin M, Dong XW, Guo XL. Role of the interaction between galectin-3 and cell adhesion molecules in cancer metastasis. Biomed Pharmacother 2015;69:179-85. [Crossref] [PubMed]
- Opiłka MN, Lorenc Z, Starzewska M, et al. Cell adhesion molecules in terms of carcinogenesis. Pol Przegl Chir 2014;86:151-7. [Crossref] [PubMed]
- Zhang XF, Pan K, Weng DS, et al. Cytotoxic T lymphocyte antigen-4 expression in esophageal carcinoma: implications for prognosis. Oncotarget 2016;7:26670-9. [Crossref] [PubMed]
- Wu J, Long Z, Cai H, et al. High expression of WISP1 in colon cancer is associated with apoptosis, invasion and poor prognosis. Oncotarget 2016;7:49834-47. [Crossref] [PubMed]
- Gröne J, Weber B, Staub E, et al. Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, -8 and -12. Int J Colorectal Dis 2007;22:651-9. [Crossref] [PubMed]
- Bujko M, Kober P, Mikula M, et al. Expression changes of cell-cell adhesion-related genes in colorectal tumors. Oncol Lett 2015;9:2463-70. [PubMed]
- Konczal M, Koteja P, Stuglik MT, et al. Accuracy of allele frequency estimation using pooled RNA-Seq. Mol Ecol Resour 2014;14:381-92. [Crossref] [PubMed]


