
Effects of GLS1 on the epithelial-mesenchymal transition of hepatocellular carcinoma in vitro and in vivo
Introduction
Hepatocellular carcinoma (HCC), as a malignant tumor, is typified by rich blood supply, rapid progression as well as strong invasion and metastasis. Induced by complex processes, HCC plays a carcinogenic role through interactions between internal factors (e.g., multi-gene participation and regulation through differential expressions of genes at various stages) and external factors (e.g., chemical toxicants and pathogens). Currently, HCC is mainly treated by traditional surgery, systemic chemotherapy, in vitro radiotherapy, radiofrequency ablation, liver transplantation together with novel strategies such as immune therapy, targeted therapy and gene therapy (1). Although great progress has been made on the diagnosis and surgical treatment of HCC in the past decades, the long-term prognosis remains poor, with the 5-year survival rate of only 33–44% and the 5-year recurrence rate of over 70%. The postoperative recurrence and poor efficacy of HCC can mainly be attributed to its strong invasion and metastasis. Therefore, it is of great significance to find a more effective therapy for reducing the invasion and metastasis capacities as well as recurrence rate of HCC so as to improve the prognosis and survival rate of patients (2,3). In recent years, new strategies such as targeted therapy and gene therapy have developed rapidly, showing therapeutic prospects.
The metabolic way of cancer cells is different from that of normal ones. Many cancer cells, which absorb a large amount of glucose, are mainly decomposed and utilized through aerobic glycolysis, also known as the Warburg effect (4,5). In addition, cancer cells undergo metabolism while being addicted to glutamine, i.e., they absorb and utilize more glutamine and exhibit high glutaminase activity (6). Besides providing a material basis for the synthesis of biological macromolecules in cancer cells, glutamine metabolism also participates in the regulation of intracellular signaling pathways and play an important role in the onset and progression of malignant tumors (7). The degradation of extracellular matrix is a crucial process during the invasion and metastasis of tumor cells. Based on the characteristics of degradation, matrix metalloproteinases (MMPs) dominantly control tumor invasion and metastasis, of which MMP2 and MMP9 are most important. Epithelial-mesenchymal transition (EMT) refers to the process during which epithelial cells transform to mesenchymal ones and obtain their phenotype under specific pathophysiological conditions (7). Through EMT, epithelial cells lose polarity, resist apoptosis and manage to degrade extracellular matrix, contacting less with surrounding cells and matrix. Meanwhile, their adhesion is reduced, whereas the migration, mobility and invasion are enhanced, accompanied by transition from epithelial phenotype into mesenchymal phenotype (8). These features are closely related to the in situ invasion and distal metastasis of malignant tumors. Therefore, HCC may be effectively treated by inhibiting glutamine metabolism, and GLS1 is a potentially new target for the treatment of human malignancies.
Methods
Cell lines and reagents
HCCLM3 cells were purchased from the FuHeng Cell Center (China). Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin GLS1 lentiviral expression vector, its empty plasmid (CD510B-1), lentiviral vector sgRNA-GLS and negative control virus sgRNA-CON244 were purchased from GeneChem (China). DMEM, trypsin and fetal bovine serum (FBS) were bought from Gibco (USA). Trypan blue, DMSO, penicillin and streptomycin were obtained from Sigma (USA). ELISA kits for MMP2 and MMP9 were purchased from SouthernBiotech (USA). GLS1 antibody was purchased from Abcam (USA) and EMT-related antibodies were bought from CST (USA). Opti-MEM, Lipofectamine TM 2000 and Trizol were bought from Invitrogen (USA).
Cell culture
HCCLM3 cells were cultured in DMEM containing 10% FBS, 1% penicillin and 1% streptomycin, and placed in an incubator at 37 °C with 5% CO2 and full humidity. The cells were passaged when the confluence reached about 85%.
Construction of cells stably overexpressing GLS1
Well-grown HCCLM3 cells were dispersed after digestion using trypsin, counted, seeded in a 6-well plate at the density of 2.5×105 cells/well and placed in an incubator at 37 °C with 5% CO2 and full humidity. Transfection was conducted when the confluence reached 80–90%. HCCLM3 cells were transfected with two lentiviral vectors (PCDH-CMV-MCS-EF1-Puro GLS1 and PCDH-CMV-MCS-EF1-Puro) respectively, dispersed after digestion using trypsin, centrifuged and resuspended 48 h later. Then a culture medium containing puromycin was added for culture and screening. As a result, a cell line stably overexpressing GLS1 was obtained and referred to as LV-GLS1. The cells transfected with PCDH-CMV-MCS-EF1-Puro empty control plasmid were used as control group (LV-Con). Afterwards, the cells were placed in an incubator and cultured for experiments below.
Construction of cells with GLS1 knockdown
Well-grown HCCLM3 cells were dispersed after digestion using trypsin, counted, seeded in a 6-well plate at the density of 2.5×105 cells/well and placed in an incubator at 37 °C with 5% CO2 and full humidity. Transfection was conducted when the confluence reached 80–90%. HCCLM3 cells were transfected with two lentiviral vectors (sgRNA-GLS and sgRNA-CON244) respectively, dispersed after digestion using trypsin, centrifuged and resuspended 48 h later. Then a culture medium containing puromycin was added for culture and screening. As a result, a cell line with GLS1 knockdown was obtained and referred to as sgRNA-GLS1. The cells transfected with sgRNA-CON244 were used as control group (sgRNA-Con). Afterwards, the cells were placed in an incubator and cultured for experiments below.
MTT assay
HCCLM3 cells were dispersed after digestion using trypsin, counted and inoculated into a 96-well plate at the final concentration of 1×105/mL (100 µL/well). MTT assay was performed to detect cell proliferation on the 1st, 2nd, 3rd and 4th days respectively by measuring the optical density at 490 nm with a microplate reader.
Transwell assay
Transwell chambers coated and uncoated with Matrigel gel were employed for cell migration and invasion experiments respectively. The cells were resuspended in serum-free DMEM, starved for 24 h, and then adjusted to a concentration of 1×105/mL. Then 200 µL of cell suspension was incubated in the upper chamber. Subsequently, 600 µL of DMEM containing 20% FBS was added to a 24-well plate, into which the Transwell chamber was placed. After 24 h of routine incubation, the Transwell chamber was taken out and washed gently with 0.01 M PBS. Then adherent cells in the chamber were gently wiped off with a cotton swab, fixed in 95% ethanol, and finally stained with crystal violet. Five visual fields (200×) under an inverted microscope were randomly selected to count cells penetrating the chamber.
Cell scratch assay
After 24 h of culture, the cells were dispersed after digestion using trypsin into a single cell suspension through repeated pipetting, with the density adjusted to 4×105 cells/mL. Then the cells were incubated in a 6-well plate, 2 mL each well. When the cell density reached approximately 80–90%, straight lines were drawn evenly on the plate using a 10 µL pipette point. After washing with PBS, the remaining cells were incubated in a culture medium containing 1% serum. Fixed scratches were photographed at 0, 24, 48 h, respectively, and the corresponding degrees of cell fusion were recorded. The experiments were repeated three times.
Western blot
The cells were dispersed after digestion using trypsin, collected by centrifugation, resuspended by adding RIPA (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), ultrasonicated and centrifuged at 12,000 rpm for 10 min at 4 °C. Total protein concentration was measured according to the instructions of BCA kit. Afterwards, 50 µg of protein from each sample was resolved on SDS-PAGE. The protein was then transferred to a nitrocellulose membrane, blocked with 5% skimmed milk for 1 h at room temperature, incubated with primary antibodies overnight at 4 °C, washed, and incubated with secondary antibodies at 37 °C for 1 h. After ECL development and scanning, relative protein expressions were analyzed by Quantity-One after internal reference correction.
ELISA
Capture antibody was diluted by 1:200, added into a 96-well plate at 100 µL/well, and coated overnight at 4 °C. After the capture antibody was shaken off, the plate was washed 4 times using 1× washing buffer (100 µL/well), 1 min each time, and blotted on absorbent paper at the last time. Then it was blocked with blocking solution (100 µL/well) at room temperature for 2 h. Subsequently, the plate was washed 4 times using 1× washing buffer (100 µL/well), 1 min each time, and blotted on absorbent paper at the last time. Standard substance was diluted to six concentrations, with one duplicate well for each concentration. The blocking solution was thereafter shaken off and the plate was washed twice. The blank well was added with 100 µL of diluent, the standard substance wells were added with those at different concentrations, and the sample well was added with 100 µL of supernatant. Detection antibody was added into each well (100 µL/well) (capture antibody was diluted by 1:200) and incubated at room temperature for 2 h. The liquid was shaken off, and the plate was washed 4 times. Then reaction solution was added (100 µL/well), incubated at room temperature for 10 min, and then sealed using sealing membrane. The liquid was shaken off, and the plate was washed 4 times again. Chromogenic reagent was added (100 µL/well), and observation was carried out within 40 min. The absorbance of each group was measured using a microplate reader, and a standard curve was plotted with a specific program. Concentrations were calculated through the standard curve based on the measured absorbance of each group.
Immunochemical assay
Sections were baked at 60 °C for 10 min, deparaffinized (washing twice by xylene, 10 min each time), and hydrated through a descending ethanol series to distilled water (washing twice by absolute ethanol, 5 min each time; 95% ethanol, 1 min; 90% ethanol, 1 min; 80% ethanol, 1 min; 70% ethanol, 1 min). Then the sections were washed three times with PBS, 2 min each time. After removal of the liquid, tissues were circled with a pap pen. The sections were then incubated with 3% H2O2 at room temperature in dark for 10 min, boiled with 200 mL of 0.01 M citrate buffer (pH 6.0) for 10 min in a microwave, and cooled down to room temperature. After removal of the liquid, the sections were washed three times with PBS, 2 min each time. Then they were incubated with primary antibody at 4 °C overnight and then with horseradish peroxidase-labeled goat anti-rabbit secondary antibody at 37 °C for 60 min, washed three times with PBS (2 min each time), and subjected to DAB color development and nuclei staining with hematoxylin for 5 min. Finally, the sections were mounted by neutral mounting medium, observed under a microscope and photographed.
Construction of in vivo tumor cells
All animal experiments were approved by the ethics committee of Affiliated Drum Tower Hospital of Nanjing Medical University (Number: DTH-20160109), and performed in compliance with corresponding guidelines. Greatest efforts were made to minimize animals’ suffering. Nude mice were subcutaneously implanted with HCCLM3-LV-GLS1 cells (Group A, GLS1 overexpression), HCCLM3-sgRNA-GLS1 cells (Group B, GLS1 knockdown) and HCCLM3 cells (Group C), respectively. The influences of GLS1 expression on HCCLM3 cell tumorigenesis, distal metastasis and tumor growth were assessed. The mice were killed five weeks after tumor formation to compare the tumor size and to observe whether metastasis occurred in the liver and lungs.
HE staining
Sections were deparaffinized (washing twice by xylene I and II, 5 min each time), and hydrated through a descending ethanol series to distilled water (washing twice with absolute ethanol, 2 min each time; 95% ethanol, 1 min; 90% ethanol, 1 min; 80% ethanol, 1 min; 70% ethanol, 1 min). Then the sections were stained by hematoxylin for 5 min, rinsed by tap water, differentiated in hydrochloric acid-ethanol for 10 s, immersed in tap water for 10 min, immersed in 75% and 85% ethanol for 2 min respectively, and stained by eosin for 2 min. Finally, the sections were dehydrated, transparentized, mounted by neutral mounting medium, and observed under the microscope.
Statistical analysis
All data were analyzed by GraphPad. All experiments were performed in triplicate, and the results are expressed as mean ± standard deviation. P<0.05 was considered statistically significant.
Results
Construction of HCCLM3 cells with GLS1 overexpression and knockdown
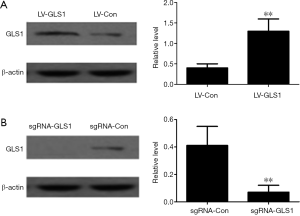
As shown in Figure 1A and Figure 1B, HCCLM3 cells with GLS1 overexpression (HCCLM3-LV-GLS1) and knockdown (HCCLM3-sgRNA-GLS1) had been successfully constructed.
Effects of GLS1 overexpression and knockdown on cell proliferation
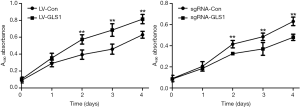
MTT assay exhibited that the proliferation of HCCLM3 cells with GLS1 overexpression (HCCLM3-LV-GLS1) was significantly promoted with extended time (P<0.01) (Figure 2A), but that of HCCLM3 cells with GLS1 knockdown (HCCLM3-sgRNA-GLS1) was significantly inhibited (P<0.01) (Figure 2B).
Effects of GLS1 overexpression and knockdown on cell migration rate
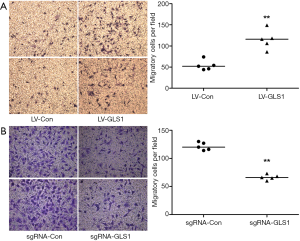
Transwell assay showed that the migration rate of HCCLM3-LV-GLS1 cells increased 24 h after transfection (Figure 3A). In contrast, the migrate rate of HCCLM3-sgRNA-GLS1 cells decreased (Figure 3B).
Effects of GLS1 overexpression and knockdown on cell invasion
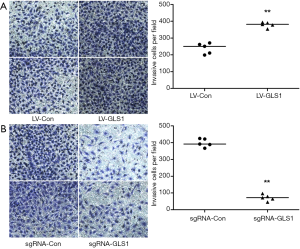
Transwell assay showed that the invasion capacity of HCCLM3-LV-GLS1 cells rose 48 h after transfection (Figure 4A), but that of HCCLM3-sgRNA-GLS1 cells reduced (Figure 4B).
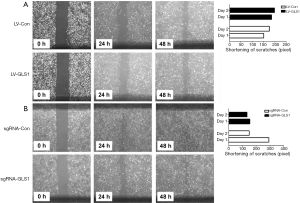
Effects of GLS1 overexpression and knockdown on healing of scratches
Scratch assay revealed that 24 and 48 h after transfection, the healing of scratches using HCCLM3-LV-GLS1 cells was accelerated (Figure 5A), whereas that of HCCLM3-sgRNA-GLS1 cells was decelerated (Figure 5B).
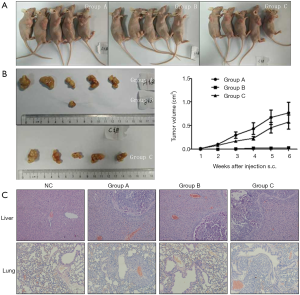
Effects of GLS1 overexpression and knockdown on subcutaneous tumors
Nude mice were subcutaneously implanted with HCCLM3-LV-GLS1 cells (Group A, GLS1 overexpression), HCCLM3-sgRNA-GLS1 cells (Group B, GLS1 knockdown) and normal HCCLM3 cells (Group C) respectively to clarify whether GLS1 expression affected HCCLM3 cell tumorigenesis and promoted or inhibited distal metastasis. After modeling, tumor growth was observed each week. The mice were killed 5 weeks after tumor formation to compare the tumor size and to observe whether metastasis occurred in the liver and lungs. Every mouse in Group A and Group C formed tumors, but only one mouse in Group B did so. Group A had a similar tumor size to that of Group C, whereas Group A and Group B had significantly different tumor sizes (Figure 6A,B). In other words, GLS1 knockdown significantly reduced the tumor formation rate and tumor volume that were barely affected by GLS1 overexpression. Pathological sections showed that tumors in situ were accompanied by pulmonary metastasis, and Group B was less prone to metastasis than the other two groups (Figure 6C).
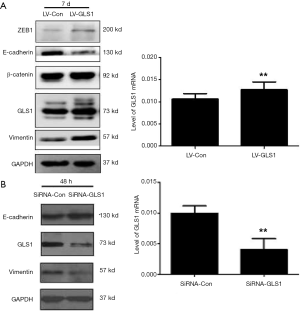
Effects of GLS1 overexpression and knockdown on EMT-related protein expressions
Western blot exhibited that the expression of epithelial marker E-cadherin decreased in HCCLM3-LV-GLS1 cells, but those of mesenchymal marker vimentin and transcription factor ZEB1 negatively regulating E-cadherin increased (Figure 7A), indicating that the EMT process of these cells was facilitated. Contrarily, such process of HCCLM3-sgRNA-GLS1 cells was suppressed (Figure 7B).
To further clarify the mechanism by which GLS1 affected tumor onset and metastasis, we detected the expressions of EMT-related proteins in the tumor-bearing mouse model.
In Group A, E-cadherin was mildly positively expressed, with a diffuse distribution. Vimentin was positively expressed mainly around tumors. Both diffusely distributed, Slug and ZEB1 were moderately and mildly positively expressed respectively (Figure 8A). With decreasing GLS1 expression, the expression of E-cadherin in Group B was up-regulated but that of vimentin was down-regulated (Figure 8B). The expressions of Slug and ZEB1 in EMT-related signaling pathways were also up-regulated. Group C had opposite outcomes to those of Group B (Figure 8C).
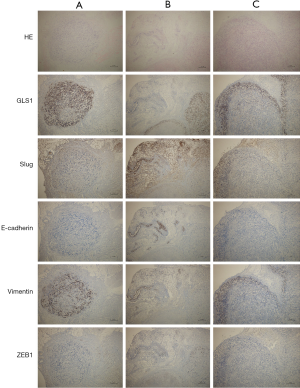
EMT occurred in a specific microenvironment, thereby initiating metastasis. Immunohistochemical assay exhibited that with increasing GLS1 expression, epithelial phenotypes in subcutaneous tumor cells were gradually replaced by mesenchymal ones, so their invasion and metastasis capacities were augmented. The in vivo results were consistent with in vitro ones.
Effects of GLS1 overexpression and knockdown on MMP2 and MMP9 expressions
ELISA revealed that the expression of MMP2 in HCCLM3-LV-GLS1 cells increased (Figure 9A) but that of HCCLM3-sgRNA-GLS1 cells decreased with extended time (Figure 9B). For the in vivo study, Group A had a higher serum MMP2 expression level than that of Group B (Figure 9C).
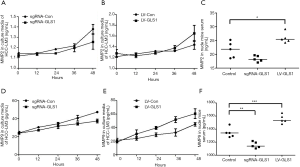
ELISA showed that the expression of MMP9 protein in HCCLM3-LV-GLS1 cells was up-regulated (Figure 9D) whereas that of HCCLM3-sgRNA-GLS1 cells was down-regulated with prolonged time (Figure 9E). As to the in vivo study, Group A had a higher serum MMP9 expression level than that of Group B (Figure 9F).
Discussion
HCC is one of the most common malignant tumors of the digestive tract, seriously endangering human health due to high and continuously increasing morbidity and mortality rates. Currently, HCC treatment is mostly limited by high invasion and metastasis capacities as well as poor prognosis. For targeted therapy and gene therapy, it is crucial to find effective interfering targets (9,10). During onset and progression, tumor cells are mainly energized via the aerobic glycolytic pathway. Besides, tumor cells depend on glutamine metabolism to provide a material basis for the synthesis of biomacromolecules in growth and proliferation. The key enzyme GLS1 in glutamine metabolism has attracted wide attention (11-13).
In this study, HCCLM3 cells with GLS1 overexpression and knockdown were constructed respectively. The migration and invasion of HCCLM3 cells, which were enhanced by GLS1 overexpression, were attenuated after knocking down GLS1. In addition, the healing of scratches using HCCLM3-LV-GLS1 cells was accelerated 24 and 48 h after transfection, whereas that of HCCLM3-sgRNA-GLS1 cells was decelerated. In HCC tissues, E-cadherin protein expression is significantly lower than those of paracancerous and normal tissues (14,15). The patients with high E-cadherin protein expression have significantly better differentiation and prognosis than those of the patients with low expression. Probably, low E-cadherin protein expression in HCC cells significantly decreases intercellular adhesion, so they are more prone to detachment from primary foci and metastasis, indicating that down-regulation of this protein facilitates metastasis. In this study, the expression of epithelial marker E-cadherin decreased in HCCLM3-LV-GLS1 cells, but those of mesenchymal marker vimentin and transcription factor ZEB1 negatively regulating E-cadherin increased. Therefore, GLS1 promoted the EMT process of HCC cells. As evidenced by in situ hybridization, PCR and immunohistochemical assay, MMP2 and MMP9 are highly expressed in colon cancer and breast cancer (16,17). Since the expressions of MMP2 and MMP9 proteins in HCCLM3-LV-GLS1 cells were elevated with extended time herein, GLS1 overexpression promoted their expressions. Moreover, in vivo experiments showed that GLS1 knockdown significantly decreased the tumor formation rate and tumor volume that were hardly affected by GLS1 overexpression. With reducing GLS1 expression, the expression of E-cadherin in Group B was up-regulated but that of vimentin was down-regulated. In the meantime, the expressions of Slug and ZEB1 in EMT-related signaling pathways were also up-regulated.
Conclusions
In summary, inhibiting glutamine metabolism is an effective strategy for HCC therapy, and GLS1 may be a novel target for treating human malignant tumors.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.14). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the ethics committee of Affiliated Drum Tower Hospital of Nanjing Medical University (Number: DTH-20160109), and performed in compliance with corresponding guidelines. for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis 2013;4:e532 [Crossref] [PubMed]
- Takahashi H, Ishii H, Nishida N, et al. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol 2011;18:1166-74. [Crossref] [PubMed]
- Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest 2013;123:3678-84. [Crossref] [PubMed]
- Glasauer A, Chandel NS. Targeting antioxidants for cancer therapy. Biochem Pharmacol 2014;92:90-101. [Crossref] [PubMed]
- Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett 2015;356:156-64. [Crossref] [PubMed]
- Huang L, Xu AM, Liu W. Transglutaminase 2 in cancer. Am J Cancer Res 2015;5:2756-76. [PubMed]
- Ge Y, Yan X, Jin Y, et al. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet 2015;11:e1005726 [Crossref] [PubMed]
- Mohamed A, Deng X, Khuri FR, et al. Altered glutamine metabolism and therapeutic opportunities for lung cancer. Clin Lung Cancer 2014;15:7-15. [Crossref] [PubMed]
- Yuan C, Liu C, Zou N. MicroRNA-340 Targets NF-κB1 to Inhibit Cell Proliferation, Migration and Invasion in Hepatocellular Carcinoma Cell Line. J Biomater Tissue Eng 2016;6:649-58. [Crossref]
- Reyes RK, Motiwala T, Jacob ST. Regulation of glucose metabolism in hepatocarcinogenesis by microRNAs. Gene Expr 2014;16:85-92. [Crossref] [PubMed]
- Phillips MM, Sheaff MT, Szlosarek PW. Targeting arginine-dependent cancers with arginine-degrading enzymes: opportunities and challenges. Cancer Res Treat 2013;45:251-62. [Crossref] [PubMed]
- Yu D, Shi X, Meng G, et al. Kidney-type glutaminase (GLS1) is a biomarker for pathologic diagnosis and prognosis of hepatocellular carcinoma. Oncotarget 2015;6:7619-31. [PubMed]
- Chen L, Cui H. Targeting Glutamine Induces Apoptosis: A Cancer Therapy Approach. Int J Mol Sci 2015;16:22830-55. [Crossref] [PubMed]
- Valencia T, Kim JY, Abu-Baker S, et al. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell 2014;26:121-35. [Crossref] [PubMed]
- Giacobbe A, Bongiorno-Borbone L, Bernassola F, et al. p63 regulates glutaminase 2 expression. Cell Cycle 2013;12:1395-405. [Crossref] [PubMed]
- Katt WP, Antonyak MA, Cerione RA. Simultaneously targeting tissue transglutaminase and kidney type glutaminase sensitizes cancer cells to acid toxicity and offers new opportunities for therapeutic intervention. Mol Pharm 2015;12:46-55. [Crossref] [PubMed]
- Oermann EK, Wu J, Guan KL, et al. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol 2012;23:370-80. [Crossref] [PubMed]


