Circular RNA shifts the miRNA binding panel and takes a role in hepatocellular carcinoma progression
Circular RNAs (circRNAs) are a novel type of RNA molecule that covalently joint its 5' and 3' end together and form a closed single strand annular. CircRNAs were first reported by Sanger et al. as a plant viroid RNA in 1976 (1). In decades, circRNAs were rarely investigated mainly because they are usually expressed at a relative low level. It is also difficult to distinguish circRNAs from the corresponding linear RNAs before the second-generation sequencing technology was invented. Until recently, by using next generation sequencing, Salzman et al. identified hundreds of back-spliced RNA products resistant to exonuclease RNase R, representing the circRNAs (2). From then on, thousands of circRNAs were reported in a variety of cells and tissue samples from human or other species (3). At first, circRNAs were generally regarded as a “by-product” of RNA splicing process and did not gain enough attention. However, more and more studies have shown that expression of circRNAs have significant tissue or cell specific manner, suggesting the underlying biological functions of circRNA (3). Many researchers focused on roles of circRNAs during varieties of biological process. At present, there are four functional models of circRNAs. Firstly, circRNA could achieve as a miRNA sponge that competitively bind endogenous miRNAs (4,5). Inner these circRNAs, there are diverse number of binding sites for one or few miRNAs, influencing their intracellular distribution or binding panel. Secondly, many studies also identify circRNA as a protein sponge that competitively bind endogenous proteins (6,7). Just as the role in miRNAs sponge, these circRNAs can bind certain proteins competitively and change their location and function. Thirdly, circRNA may function as a protein scaffolding in some protein complexes (8,9). Different from protein sponge, the function of circRNAs serving as a scaffold is not playing a role in regulating protein functions, but providing a site for protein interaction and forming larger functional complexes. Lastly circRNA could be considered as a coding gene that directly expresses a peptide or protein (10,11). These circRNAs directly translate functional peptides or proteins like classical mRNAs but in a different manner by utilizing internal ribosomal entry site or m6A modification. Among these functional models, miRNA sponge is the most frequently studied one. A typical case of miRNA sponge is the antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as) gene. In general, the final transcriptional product of CDR1as gene is a circular RNA. As is reported, CDR1as have 74 miR-7 binding sites (5). CDR1as located in the cytoplasm can specifically absorb miR-7 molecules, then the miR-7 downstream genes are released (5). In this model, level of miRNA sponge molecular can subtly control the binding panel of target miRNAs. Except for miR-7 binding sites, there is also a site of miR-671. When miR-671 binds to CDR1as, pathways that AGO2 mediate RNA interference will be activated and CDR1as will be degraded. Thus, the absorbed miR-7 will be released to regulate downstream genes (4). Other than CDR1as, there are many researches focused on the miRNA sponge circRNAs. As an interesting case, Han et al. reported that circ-MTO1 can directly bind miR-9, resulting in upregulation of p21 and suppressing hepatocellular carcinoma (HCC) progression (12). This research gives us a solid evidence that circRNAs work as miRNA sponge and gives some new points for HCC molecular pathology.
HCC is the major type of primary liver cancer, which arising from hepatocytes (13). Besides, HCC is the third leading cause of cancer-related death, with the 5 years survival rate low as 6.9% and with the overall median survival time of only 11 months (14). HCC becomes one of the thorny clinical challenges due to its high clinical incidence, difficultly of early diagnosis and quick relapse after surgery. Major epidemiological risk factors of HCC include hepatitis B virus, hepatitis C virus, aflatoxin B1-contaminated food, alcohol, and non-alcoholic liver diseases (14). Benefit from the development of next generation sequencing technology, exploration of molecular pathogenesis of HCC is greatly improved. In general, both genetic and epigenetic alterations are contribute to the HCC progression (15). Genetic changes include high expression of some oncogenes like MET, MYC, TERT, CCND1, FGF19, low expression of some tumor suppressor genes such as TP53, CDKN2A, CDKN2B, somatic mutations in protein coding genes including CTNNB1, TP53, promoter mutations leading to enhanced gene expression like TERT gene, and virus genome integration that leads to disturbance of endogenous regulatory genes (15). Epigenetic alterations include DNA or histones modification in specific genes, chromosome remodeling, noncoding RNAs as well as microRNAs that globally influence many downstream genes (15). These changes lead to alteration of transcriptome and signal network, and promote HCC carcinogenesis at last (15). Although there exist several researches considering the several genetic changes the mechanism of molecular pathogenesis of HCC is still elusive.
MiRNAs is a class of small intracellular RNAs about 20–24 nucleotides length with regulatory functions. MiRNAs play important roles in HCC occurrence, development and metastasis (16). Role of miR-9 in HCC remains unclear, although some researchers tend to support miR-9 play an important part in oncogenesis. Tan et al. reported that miR-9 is one of the most highly expressed miRNAs in SK-Hep-1 cells which could promote expression of E-cadherin (17). Sun et al. found that upregulation of miR-9 can increase migration and invasion of HCC through KLF17 (18). Liu et al. found that miR-9 can directly decrease the expression of N-acetylgalactosaminyltransferase 4 (GALNT-4) and promote HCC migration, invasion, anoikis resistance and stemness maintenance (19). Cui et al. reported that in hepatocytes, miR-9-RXRA pathway mediates the lipid metabolism transformation and promotes cell proliferation (20).
Here we mainly show one research that confirmed the relation between HCC and miR-9. The first study was conducted by Han et al. reporting circ-MTO1 can directly bind to miR-9 and suppress HCC progression (12). In their work, they used RNA in vivo precipitation (RIP) to select miRNAs directly binding to circ-MTO1 and 20 miRNAs were found finally. In addition, they reversely conform these binding miRNAs by circ-MTO1 pull-down experiments, and found specific enrichment of miR-9 but no other miRNAs (12). Furthermore, the interaction of miR-9 and circ-MTO1 also verified by fluorescence in situ hybridization (FISH). MiR-9 upregulation in HCC cell line SK-Hep1 (17) was also verified by Tan et al. After using both the artificial overexpression and inhibition of miR-9, they identified the circ-MTO1/miR-9/p21 axis in HCC progression (12). Additionally, cir-MTO1 is usually downregulated in HCC and has negative relationship with the overall survival rate of HCC patients (12). Reduction of circ-MTO1 leads to release of miR-9, which suppress p21 expression and promote cell proliferation (12). Using circRIP, this study presented us a strong evidence about interaction between miR-9 and circ-MTO1, and strengthened the importance of both miR-9 and circ-MTO1 in molecular pathogenesis mechanism of HCC. However, it is regrettable that we could not witness the circ-MTO1 knock-out model to conform the function of circ-MTO1 in vivo. Maybe without this deficiency, this work would have published in journals of higher level. Certainly, this problem is also ubiquitous for other current circRNA researches beyond this study, as knocking out circRNA usually together diminished its corresponding linear mRNA, and further rescue experiment are certainly required.
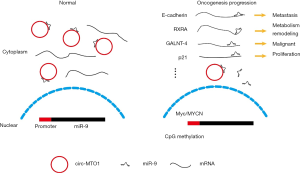
Summarizing all relevant studies, we can get a functional model of circ-MTO1 and miR-9 in HCC, showed in Figure 1. Normal hepatocytes have high expression of circ-MTO1. MiR-9 could competitively bind to circ-MTO1, then the downstream genes were all normally expressed, including E-Cadherin, RXRA, GALNT4, p21, etc. Accompany with HCC progression, circ-MTO1 reduction caused miR-9 releasing and inhibition of downstream genes, facilitating some important oncogenesis behavior like cell metastasis, uncontrolled proliferation, metabolism transformation and so on. In this model, changing of circ-MTO1 expressing level altered the concentration of functional miR-9 and shifts binding panel of miR-9. This study provided us a strong evidence of circ-MTO1 competitively binding miR-9 and shifting the binding panel of miR-9, and showed some useful information for both diagnosis and treatment for HCC in the future. This study revealed a new evidence of miRNA sponge model of circRNAs, that may greatly promote functional study about circRNAs.
Acknowledgments
Funding: National Natural Science Foundation of China (No. 81370072, 81572477 and 81772683).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14:361-9. [Crossref] [PubMed]
- Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 2017;24:357-70. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110. [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Ozakyol A. Global Epidemiology of Hepatocellular Carcinoma (HCC Epidemiology). J Gastrointest Cancer 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Ho DW, Lo RC, Chan LK, et al. Molecular Pathogenesis of Hepatocellular Carcinoma. Liver Cancer 2016;5:290-302. [Crossref] [PubMed]
- Xu J, Li J, Zheng TH, et al. MicroRNAs in the Occurrence and Development of Primary Hepatocellular Carcinoma. Adv Clin Exp Med 2016;25:971-5. [Crossref] [PubMed]
- Tan HX, Wang Q, Chen LZ, et al. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med Oncol 2010;27:654-60. [Crossref] [PubMed]
- Sun Z, Han Q, Zhou N, et al. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol 2013;7:884-94. [Crossref] [PubMed]
- Liu Y, Liu H, Yang L, et al. Loss of N-Acetylgalactosaminyltransferase-4 Orchestrates Oncogenic MicroRNA-9 in Hepatocellular Carcinoma. J Biol Chem 2017;292:3186-200. [Crossref] [PubMed]
- Cui M, Xiao Z, Wang Y, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res 2015;75:846-57. [Crossref] [PubMed]