
A novel window opened: EBV-driven enhancer-promoter loops in lymphocytic immortalization
Epstein-Barr virus (EBV) is one of the human herpesvirus, which infects more than 90% of the global population (1). EBV infection causes the disorder of the immune system and has an essential role in the development of cancers (2). EBV mainly infects CD21 receptor-positive B lymphocytes in vivo (3). Thus, EBV is considered as a B lymphotropic virus (4). In primary EBV infection, EBV resides as a slow-cycling phenotype in the long-lived memory B lymphocytes of infected individuals and cannot cause major diseases because of the long-term latent capability (5). Besides, EBV is also able to transform B cells into continuously proliferating lymphoblastoid cell lines (LCLs) in vitro (6). However, the underlying mechanisms responsible for the relationships between EBV and B lymphocytes not completely understood.
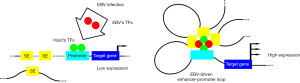
In the cellular nucleus, diverse activity of transcription-factor-bound enhancers makes the dynamic and restricted control of transcriptional network (7). In 2013, a study revealed that transcription factors form with large enhancer domains in some genomic sites (also known as super-enhancers) initiating genes that are important for the pluripotent state in embryonic stem cells (8). The super-enhancer was composed of multiple regions in the genome and highly levels of binding transcription factors. It could loop to target genes and activating transcription (9). Recent studies have suggested that the large transcription complexes regulate a series of biological functions like cell proliferation, mutation and drug sensitivity in cancers (8-10). A recent study has demonstrated that the convergent actions of EBV transcription factors, NF-kB subunits and other host cofactors lead to the formation of EBV super-enhancers, which mediates the host oncogene expression in lymphoblastoid cells (11). Moreover, enhancer RNAs were activated by EBNA2, which have been verified as a component of EBV super-enhancers, which also plays a critical role in lymphoblastoid cell growth and survival (12). Above all, these evidences suggested that the viral proteins were strongly associated with host cells immortalization. However, the mechanisms responsible for the virus infection in activating host cells via the enhancer-promoter loops are still unclear.
In a recently published study, Jiang et al. (13) have established a 3D chromatin map of the EBV-infected LCLs. Using the RNAPII ChIA-PET technique and related data analysis, the authors identified about thirty percent of genes associated with proliferation in LCLs were linked to viral enhancers. Jiang and his colleagues focused on the direct target genes of EBV subsequently and verified the consequences. They showed that MYC-EBV super-enhancers were essential for MYC expression and cell growth by deleting the MYC locus using CRISPR/Cas9 technique. The authors also confirmed that EBV transcription factors EBNA3A/3C altered CDKN2A/B spatial organization to suppress senescence, and that EZH2 inhibition reduced the looping at the CDKN2A/B loci and decreased LCLs growth. These findings provide a deep insight of the spatial organization during the transformation of B lymphocytes by EBV.
In summary, Jiang et al. have established EBV-related regulome in LCLs and demonstrated that hundreds of genomic sites are linked to EBV enhancers that are crucial for LCLs growth and survival (Figure 1). Besides, this study provided powerful evidence to support that virus had able to selectively target host genes which essential for their achievement of latent infection. Life activity in the cell is accurately regulated. Traditional basic biology research is based on linear (one- or two-dimensional) studies such as PCR or WB. Following the Bioinformatics, the combination of traditional methods and sequencing technology has led the laboratory research into the high-throughput, electronic data-based research. In recent years, being the beneficiary of advances in ChIP-seq technologies, an increasing number of super-enhancers will be discovered and analyzed. It brings our studies on life science into a three-dimensional research level. These underlying mechanisms of EBV enhancer-promoter loop will be crucial for antiviral drugs. However, because of the complexity of viral infection, it still has a long way to be done in order to solve this problem and accordingly to convert scientific research into clinical application. Therefore, achieving a greater understanding in virus-host interactions is fundamental. With all efforts, targeted therapy may become a promising therapeutic strategy for treating EBV associated disease in the near future.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundations of China (No. 31670171, 81728011).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.28). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chan KC, Woo JK, King A, et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 2017;377:513-22. [Crossref] [PubMed]
- Yu H, Lu J, Zuo L, et al. Epstein-Barr virus downregulates microRNA 203 through the oncoprotein latent membrane protein 1: a contribution to increased tumor incidence in epithelial cells. J Virol 2012;86:3088-99. [Crossref] [PubMed]
- Zuo L, Yu H, Liu L, et al. The copy number of Epstein-Barr virus latent genome correlates with the oncogenicity by the activation level of LMP1 and NF-κB. Oncotarget 2015;6:41033-44. [Crossref] [PubMed]
- Ou C, Sun Z, Zhang H, et al. SPLUNC1 reduces the inflammatory response of nasopharyngeal carcinoma cells infected with the EB virus by inhibiting the TLR9/NF-κB pathway. Oncol Rep 2015;33:2779-88. [Crossref] [PubMed]
- Lu Y, Qin Z, Wang J, et al. Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J Innate Immun 2017;9:574-86. [Crossref] [PubMed]
- Liu L, Zhou Q, Xie Y, et al. Extracellular vesicles: novel vehicles in herpesvirus infection. Virol Sin 2017;32:349-56. [Crossref] [PubMed]
- Bian J, Zheng J, Li S, et al. Sequential Differentiation of Embryonic Stem Cells into Neural Epithelial-Like Stem Cells and Oligodendrocyte Progenitor Cells. PLoS One 2016;11:e0155227 [Crossref] [PubMed]
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307-19. [Crossref] [PubMed]
- Pott S, Lieb JD. What are super-enhancers? Nat Genet 2015;47:8-12. [Crossref] [PubMed]
- Yuan J, Jiang YY, Mayakonda A, et al. Super-Enhancers Promote Transcriptional Dysregulation in Nasopharyngeal Carcinoma. Cancer Res 2017;77:6614-26. [Crossref] [PubMed]
- Zhou H, Schmidt SC, Jiang S, et al. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 2015;17:205-16. [Crossref] [PubMed]
- Liang J, Zhou H, Gerdt C, et al. Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc Natl Acad Sci U S A 2016;113:14121-6. [Crossref] [PubMed]
- Jiang S, Zhou H, Liang J, et al. The Epstein-Barr Virus Regulome in Lymphoblastoid Cells. Cell Host Microbe 2017;22:561-73.e4. [Crossref] [PubMed]


