
MiR-875-5p inhibits hepatocellular carcinoma cell proliferation and migration by repressing astrocyte elevated gene-1 (AEG-1) expression
Introduction
Liver cancer or hepatocellular carcinoma (HCC) is the second highest worldwide cancer with high mortality rate but limited therapeutic options (1,2). HCC is influenced by various factors, including hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol and nonalcoholic fatty liver disease (NAFLD) (3,4). However, the pathogenesis of HCC is not yet completely known.
Metastasis is the typical characteristics of cancers, consisting of tumor invasion, tumor cell dissemination, colonization of distant organs and metastatic outgrowth, which is one of the greatest challenges for cancer therapy (5). Effective prevention of metastasis would be the critical step for cancer therapy and prolonging lifespan. However, until now we still lack effective clinical therapy to inhibit metastasis (6).
Astrocyte elevated gene-1 (AEG-1), also known as metadherin (MTDH) has been reported to play a key role in the progression, invasion and metastasis of multiple human cancers. AEG-1 overexpression in HCC cells increases the production of angiogenic factors, such as vascular endothelial growth factor (VEGF), placental growth factor, and fibroblast growth factor α. However, the regulatory mechanism is unclear (7).
MicroRNAs are small noncoding RNAs with approximately 19–25 nucleotides in length, which can bind to the 3'-untranslated region (3'-UTR) of target genes. The binding of miRNAs with the target genes 3'-UTRs results in the cleavage of target mRNAs or inhibiting protein translation, and negatively regulates the expression of target genes (8). Increasing number of reports have shown that miRNAs are involved in many biological processes, including cell proliferation, differentiation, metastasis, apoptosis and immune responses (9). MiR-21 promotes the growth of the breast cancer cell line MCF-7 both in vitro and in vivo (10). MiR-126 restoration reduces the growth and proliferation of tumors, and miR-335 inhibits metastatic cell invasion (11). MiR-29 is proved to inhibit metastasis through the targeting of TET1 in HCC carcinogenesis and progression (12). MiR-125a-5p inhibits cell proliferation and induces apoptosis in HBV-related HCC by down-regulation of ErbB3 (13).
Although substantial efforts have been made to identify miRNA functions, the molecular mechanism of miRNAs in tumorigenesis and metastasis is still not well understood.
MiR-875-5p as a novel microRNA reported recently is found to be down-regulated in prostate cancer tissues, which impairs metastasis in prostate cancer models by repressing epithelial-mesenchymal transition (EMT) and enhances radiation response of prostate cancer cells via EGFR suppression (14). In addition, miR-875-5p works as a tumor suppressor by the down-regulation of EGFR in colorectal carcinoma (15). These results suggest the tumor-suppressive function of miR-875-5p and its role as an attractive target candidate for cancer therapy.
At present, the reports of miR-875-5p are limited and the role of miR-875-5p in HCC still remains ambiguous. Elucidation of miR-875-5p function in HCC and identification of its direct target may promote the development of HCC therapy. In the present study, we investigated the effects of miR-875-5p on MHCC97H cells by targeting AEG-1. Our data revealed that miR-875-5p is down-regulated in the HCC tissues compared with the matching paracarcinoma tissues (that is the matched adjacent non-cancer tissues). We also confirmed that miR-875-5p negatively regulates cell proliferation, migration, invasion and angiogenesis by targeting AEG-1 in vitro. These data revealed the underlying mechanism by which miR-875-5p inhibited the migration and invasion of HCC and regard miR-875-5p as a novel prognostic biomarker for HCC patientS
Methods
Patient samples and cell line
Informed consent was obtained from all the patients and the present study was approved by the Ethics Committee of the Capital Medical University in accordance with the Declaration of General hospital of Chinese PLA. Samples (including 29 pairs of HCC and the corresponding adjacent normal tissues) were collected from hepatoma patients undergoing partial hepatectomy. The hepatoma cell lines MHCC97-H was cultured and maintained in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum (FBS, Gibco, USA), 100 U/mL penicillin and 0.1 mg/mL streptomycin at 37 °C with 5% CO2.
Plasmids, miRNA, siRNA and transient transfection
The 3'-UTR or the mutant 3'-UTR of AEG-1 was cloned into the luciferase reporter vector to construct the plasmids pLUC-AEG-1 and pLUC-mut-AEG-1. The miR-875-5p mimics, siRNAs of AEG-1 and their negative controls (NC) were purchased from Ribo Biotech (Guangzhou, China). The sequences of primers used are described as followings: shAEG: AAAAGCCATCTGTAATCTTATCACTCGAGTGATAAGATTACAGATGGC. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA extraction and qRT-PCR
For clinical samples and cultured cell lines, total RNA was extracted by Trizol (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocols. The expression level of miR-875-5p was determined by qRT-PCR using the SYBR Premix Ex Taq Kit (Takara, Japan) in a DNA Engine Opticon2 system (Bio-Rad, USA). Primers were bought from Ribo Biotech. The steps for amplification were: denaturation at 95 °C for 3 min, amplification for 40 cycles at 95 °C for 12 s and 62 °C for 40 s. The melting curve was plotted from 62 to 95 °C, and read every 0.2 °C with a 2 s’ hold). The U6 small nuclear RNA was used as an internal control. The results were calculated by the 2−ΔΔCT method and represented as fold changes.
Transwell migration and invasion assay
For migration assay, MHCC97-H cells were seeded into the upper chamber of a transwell insert (pore size 8 µm, Costar) in 100 µL of serum-free medium. Medium of 600 µL containing 10% FBS was added in the lower chamber to act as a chemoattractant. Unmigrated cells were removed from the upper chamber by scraping with a cotton bud. The cells on the lower surface of the insert were fixed with 4% formaldehyde (Sigma, USA) and stained by crystal violet. For invasion assay, the upper chamber was coated by matrigel (BD, USA) according to the manufacturer’s instructions. Cells were seeded into the matrigel-coated chamber and cultured for about 48 h. The next steps were as same as the migration assay.
Immunofluorescence
MHCC97-H cells cultured on cover slides in 24-well plates were transfected with miR-875-5p mimics, siRNA of AEG-1 and their NCs. After 48 h of culture, the cells were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100 and blocked with 2% bovine serum albumin (BSA, Sigma, USA) for 1 h at room temperature. Then the cells were incubated with the primary antibodies E-cadherin and Vimentin (Abcam, USA) overnight, respectively. Subsequently, the secondary antibodies conjugated with Alexa Fluor 488 were applied. Cell nucleus was stained with DAPI (Roche, USA). The cells were visualized using a fluorescence microscope (Nikon, Japan).
Cell proliferation assay
MHCC97-H cells were seeded into a 96-well plate at a density of 5×103 cells/well. Next day, the cells were transfected with the indicated miRNAs or siRNAs. About 72 h after transfection, the cells were incubated with 10% CCK8 reagent (Dojindo, Japan) for 1 h at 37 °C. The result was detected by the automatic spectrometer (EnSpire Multimode Plate Reader, Perkin Elmer, USA) at 450 nm.
Cell cycle analysis
Cells cultured in 6-well plates were transfected with the indicated miRNAs or siRNAs. After 48 h, the cells were collected and fixed in 70% ethanol at 4 °C for 2 h. The cells were then washed and resuspended in 100 mL phosphate-buffered saline (PBS). RNase (50 mg/mL) was added for RNA removal at 37 °C for 30 min. The cells were stained with 200 mL propidium iodide (PI) solution (KeyGen Biotech, Nanjing, China) at 4 °C for 30 min before analyzing using a FACS Aria cell sorting system (BD Biosciences, San Jose, CA, USA).
HUVEC tube formation assay
HUVEC cells were seeded in a 96-well matrigel-coated plate at a density of 3×104 cells/well. miRNA/siRNA or NC were transfected into MHCC97-H cells. The supernatants were collected at 72 h after transfection to add to the HUVEC cells, respectively. The tubule elongation and branching formation were analyzed afterwards. Visualize the cells using a light microscope. Take pictures of the images of the capillary network and count the tube lengths using the Scion image software. Visualize the cells using the light microscope. Take pictures of images of the capillary network and count the tube lengths and branches using the scion image software.
Luciferase assay
The reporter plasmid pLUC-AEG-1 or pLUC-mut-AEG-1 was transfected into MHCC97-H cells with miR-875-5p mimics or the NC. After 24 h of culture, luciferase assays were performed. At least three independent experiments were performed.
Western blot analysis
After 48 h of transfection with the indicated miRNAs and siRNAs, the MHCC97-H cells were harvested for protein extraction. Proteins in lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes. After blocking, the membranes were incubated with the primary antibody (mouse anti-AEG-1, Protein Tech, Chicago, IL, USA) at 4 °C overnight and then with the secondary antibody (goat anti-mouse IgG-HRP, Santa Cruz, CA, USA) at room temperature for 1 h. Band signals were visualized using an enhanced chemiluminescence kit (Pierce, Minneapolis, MN, USA). β-actin (Santa Cruz, CA, USA) was used as the loading control.
Immunohistochemistry
Tumor samples were fixed in 10% buffered formalin, transferred to 70% ethanol and then embedded in paraffin. The paraffin lumps were sectioned and mounted on glass slides. Immunohistochemistry was performed using AEG-1 primary antibody at 4 °C overnight and then with the secondary antibody (goat anti-mouse IgG-HRP) followed by DAB staining.
Statistical analysis
All results were expressed as mean ± SD derived from at least three independent experimentS Statistical comparisons were made using an unpaired two-tailed student t-test. A P value <0.05 was considered as significant. * indicates P<0.05; ** indicates P<0.01.
Results
Decreased expression of miR-875-5p in HCC tissues
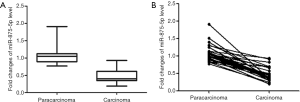
The qRT-PCR method was performed to determine the expression levels of miR-875-5p in carcinoma tissues and matched adjacent non-cancer tissues. As shown in Figure 1A, the expression levels of miR-875-5p in HCC tissues was significantly decreased compared with those in the matched adjacent non-cancer tissues (29 pairs, P<0.01). Pair-wise comparison also indicated that the expression level of miR-875-5p in carcinoma tissues was much lower than the corresponding adjacent normal tissues (Figure 1B).
MiR-875-5p inhibits metastatic traits in HCC cells
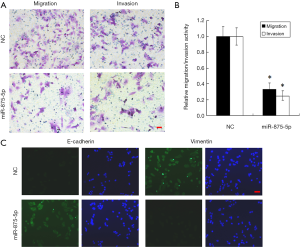
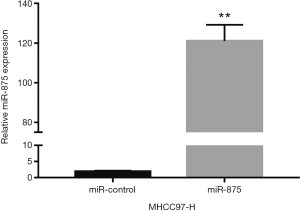
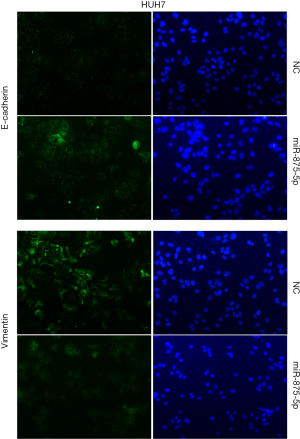
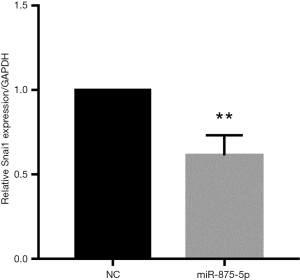
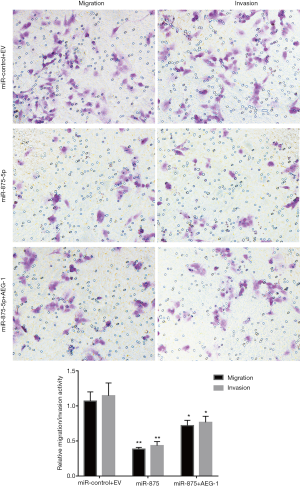
To explore the possible role of miR-875-5p in HCC cells, we first identified the effects of miR-875-5p on cancer cell migration and invasion by transwell assay. MiR-875-5p was overexpressed by transfection with miR-875-5p mimics in MHCC97-H cells indicated in Figure S1. As shown in Figure 2A,B, overexpression of miR-875-5p decreased the migratory ability of the cells with about 65% reduction compared to the NC. Similarly, the invasive capability of the cells was also significantly attenuated by 75%. EMT is the early event in tumor invasion and metastasis. Here, immunofluorescence analysis indicated that transfection of miR-875-5p mimics induced the up-regulated expression of an epithelial marker (E-cadherin) and the reduced expression of a mesenchymal marker (Vimentin), respectively (Figure 2C). The experiments were also performed in the HUH7 cells (Figure S2). In addition, another EMT marker snai1 expression was obviously reduced in miR-875-5p-overexpressed cells as determined by qRT-PCR (Figure S3). These results demonstrate that miR-875-5p inhibits metastatic traits in HCC cells.
The effects of miR-875-5p on cell proliferation and cell cycle of HCC cells
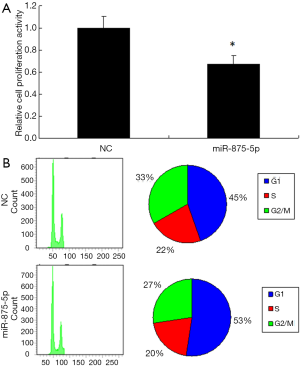
To further identify the role of miR-875-5p, we analyzed its effects on cell proliferation and cell cycle. The results showed that overexpression of miR-875-5p suppressed the cell proliferation by approximately 30% (Figure 3A). The change of cell proliferation is often due to the variation of cell cycle. Therefore, cell cycle analysis was performed by FACS after miR-875-5p transfection, showing the increased proportion of the cells in G1 phase but decreased proportion in G2/M phase compared with the control (Figure 3B). These findings indicate that miR-875-5p negatively regulates cell proliferation of HCC cells.
MiR-875-5p inhibits angiogenesis modeled by HUVEC cells in vitro
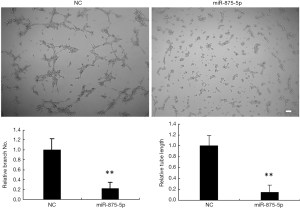
To investigate whether miR-875-5p has effects on angiogenesis, we collected the supernatant 72 h after miR-875-5p mimic control transfected into MHCC97-H cells to add to the HUVEC cells, respectively. HUVEC cells were seeded onto the matrigel-coated plates without angiogenic stimuli in advance. The tubule elongation and branching formation were analyzed 2 days culture afterwards. Visualize the cells using a light microscope. Take pictures of the images of the capillary network and count the tube lengths using the Scion image software. As shown in Figure 4, miR875-5p over-expression significantly inhibited branching formation and tubule elongation compared to control.
MiR-875-5p targets the 3'-UTR of AEG-1 to reduce AEG-1 expression
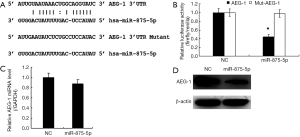
To explore the molecular mechanism of miR-875-5p, we predicted the possible target gene of miR-875-5p using the miRNA.org algorithm. As shown in Figure 5A, the binding of miR-875-5p with the 3'-UTR of AEG-1 was predicted, suggesting that AEG-1 gene may be a target of miR-875-5p. To verify the prediction, we cloned the 3'-UTR of AEG-1 into the luciferase reporter vector, and the control vector with the mutant 3'-UTR of AEG-1 was also constructed. After co-transfection of the vector with miR-875-5p mimics into MHCC97-H cells, the luciferase activity was measured, showing about 55% reduction compared with the NC group. When the binding site of miR-875-5p in the 3'-UTR of AEG-1 was mutated, the luciferase activity was rescued to the similar level with the control (Figure 5B). Furthermore, the endogenous mRNA and protein levels of AEG-1 in MHCC97-H cells were both decreased after miR-875-5p mimic transfection confirmed by qRT-PCR and Western blot assays, respectively (Figure 5C,D). Moreover, the rescue assay had been performed to provide stronger evidence that miR-875-5p suppresses HCC proliferation and migration by inhibiting the expression of AEG-1 as shown in Figure S4. Taken together, these results demonstrate that miR-875-5p targets the 3'-UTR of AEG-1 to negatively regulate AEG-1 expression.
Silencing of AEG-1 inhibits metastatic traits in HCC cells
The results above have demonstrated that miR-875-5p suppresses the metastatic traits in HCC cells and targets AEG-1 gene to reduce its expression. To further verify these findings, we checked the performance of the HCC cells (MHCC97-H) after AEG-1 knockdown using transwell migration and invasion. The results showed that knockdown of AEG-1 decreased the number of migratory and invasive cells according to the crystal violet staining, with about 55% and 65% reduction in the migration and invasion activities (Figure 6A,B).
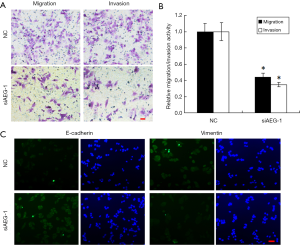
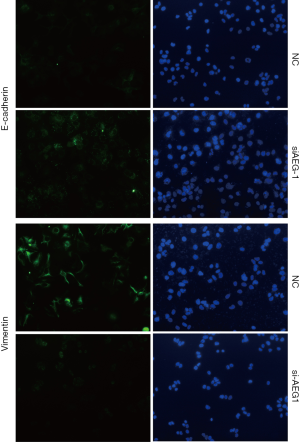
Furthermore, we also detected the effects of AEG-1 knockdown on EMT both in MHCC97-H and HUH7 cells. The results showed that AEG-1 knockdown decreased the expression of Vimentin and increased the expression of E-cadherin compared with the control analyzed by immunofluorescence analysis (Figure 6C and Figure S5). Taken together, these results demonstrate that the silencing of AEG-1 inhibits metastatic traits in HCC cells.
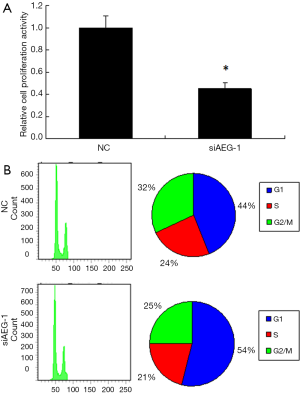
The effects of AEG-1 knockdown on cell proliferation and cell cycle of HCC cells
We further determined the effects of AEG-1 knockdown on cell proliferation and cell cycle. According to the CCK8 assay result, the cell proliferation was decreased by approximately 45% (Figure 7A). To check the variation of cell cycle, FACS analysis of the cells after PI staining was performed. The proportion of the cells in G1 phase was obviously increased, while that in G2/M phase was decreased to some extent compared with control (Figure 7B). These findings indicate that knockdown of AEG-1 is helpful to repress the cell proliferation of HCC cells and further confirm the function of miR-875-5p targeting AEG-1.
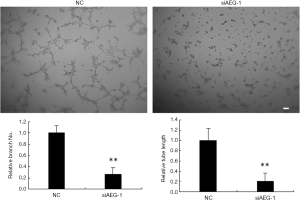
The effects of AEG-1 knockdown on angiogenesis in vitro analyzed using HUVEC cells
To investigate whether AEG-1 has effects on angiogenesis, we analyzed the branching formation and tubule elongation in vitro using HUVEC cells. Supernatants were collected at 72 h after AEG-1 siRNA or control transfected into MHCC97-H cells to add to the HUVEC cells, respectively. HUVEC cells were seeded onto the matrigel-coated plates without angiogenic stimuli ahead of time. The tubule elongation and branching formation were analyzed 2 days culture afterwards. As shown in Figure 8, knockdown of AEG-1 significantly inhibited the number of branching and the length of tubule compared to control, consistent with the effects of miR-875-5p over-expression, which further confirms that AEG-1 is the target of miR-875-5p.
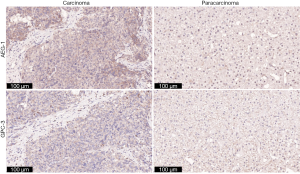
The expression of AEG-1 in HCC tissues was elevated
According to the results above, the levels of miR-875-5p are decreased in HCC tissues, and AEG-1 is a downstream target of miR-875-5p. Here, the expression of AEG-1 in clinical tissues was further determined by immunohistochemical analysis. It was found that the expression of AEG-1 in HCC tissues was relative higher than that in paracarcinoma tissues (Figure 9). Meanwhile, we also detected the expression of Glypican-3 (GPC-3), a marker for HCC. The level of GPC-3 in carcinoma tissues was higher than those in paracarcinoma tissues, similar with the trends of AEG-1. These results indicate that HCC tissues express elevated level of AEG-1.
Discussion
HCC is a worldwide malignancy with no ideal targeted therapies. To achieve permanent cure of HCC, massive efforts are urgently needed to define the mechanisms of hepatocarcinogenesis, identify solid diagnostic biomarkers and develop effective therapeutic strategies (16,17).
Recent studies have reported that MiR-875-5p exerts tumor suppressor function by repressing EMT and downregulating EGFR in prostate cancer and colorectal carcinoma tissues, showing great potential as a target candidate for cancer therapy (14,15). However, the regulatory mechanism of miR-875-5p in HCCs largely unknown. In this study, we demonstrated that miR-875-5p was obviously decreased in HCC tissues compared with the matching adjacent non-tumor tissues (Figure 1). The expression patterns of miR-875-5p in various tissues may be different. Few studies indicated that the expression of miR-875-5p was up-regulated during tooth morphogenesis (18), suggesting that miR-875-5p may also be involved in other biological processes. Furthermore, over-expression of miR-875-5p negatively regulates the proliferation, migration and invasion of HCC cells, as well as angiogenesis in vitro modeled by HUVEC cells (Figures 2-4), which triggers an imagination that miR-875-5p may be a promising therapeutic target for HCC treatment. Similarly, the enforced expression of miR-199a in HCC cells leads to cell cycle arrest at G1 phase and reduces invasive capability (19). MiR-122 reduces migration and invasion in vitro, tumorigenesis and angiogenesis in vivo, and inhibited the local invasion in the liver of nude mice (20). In contrast, MiR-221 over-expression in HCC cells increases cell proliferation, migration and invasion capability, and enhances tumorigenesis in implanted mice (21,22)
The concrete promoting or suppressing activity of miRNAs in tumorigenesis largely depends on its downstream targetS In this study, we demonstrate that miR-875-5p could target the 3'-UTR of AEG-1 gene to down-regulate its expression (Figure 5). AEG-1 is a potentially crucial mediator of tumor malignancy and a key converging point of a complex network of oncogenic signaling pathways reported in recent years (23). Similar with the expression pattern in HCC (Figure 9), the levels of AEG-1 are found to be elevated in breast carcinoma, melanoma and malignant glioma cell lines compared to their normal cellular counterparts (16). Here, the experimental results exhibited that AEG-1 knockdown exerted the similar effects with miR-875-5p overexpression, with the reduced cell proliferation, migration, invasion and angiogenesis in vitro, and cell cycle arrest at G1 phase (Figures 6-8). Similarly, inhibition of AEG-1 by siRNA significantly suppressed migration and invasion of malignant glioma cells and prostate cancer cells and lung metastasis of breast cancer cells in vivo (24).
A specific mRNA can be coordinately suppressed by several different miRNAs, and in turn one miRNA may have several targets (25). For example, except targeting AEG-1 showed in this study, miR-875-5p can also target the 3'-UTR of EGFR mRNA in CRC cells (14). On the other hand, AEG-1 can also be targeted by miR-375 in HCC cells (16). Therefore, the function networks of miRNAs and their targets are complex, and fully understanding of them is required.
Taken together, we give the evidence that miR-875-5p shows decreased levels in the HCC tissues and functions as a negative regulator in proliferation, migration, invasion and angiogenesis in vitro by targeting AEG-1. These results provide novel insights into the roles of miR-875-5p and its targets in HCC, and suggest an attractive direction for HCC target therapy.
Acknowledgments
Funding: This work was financially supported by the Beijing Natural Science Foundation [7174314], National Natural Science Foundation of China [81472328], National Science and Technology Support Program (2015BAI02B00), Capital Clinical Characteristics (Z171100001017063), Beijing municipal hospital scientific research project (PX2018057).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.22). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all the patients and the present study was approved by the Ethics Committee of the Capital Medical University in accordance with the Declaration of General hospital of Chinese PLA.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41. e23.
- Sia D, Villanueva A, Friedman SL, et al. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017;152:745-61. [Crossref] [PubMed]
- Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog 2017;16:1. [Crossref] [PubMed]
- Yu MW, Lin CL, Liu CJ, et al. Influence of Metabolic Risk Factors on Risk of Hepatocellular Carcinoma and Liver-Related Death in Men With Chronic Hepatitis B: A Large Cohort Study. Gastroenterology 2017;153:1006-17.e5. [Crossref] [PubMed]
- Massagué J, Batlle E, Gomis RR. Understanding the molecular mechanisms driving metastasis. Mol Oncol 2017;11:3-4. [Crossref] [PubMed]
- Sadhu SS, Wang S, Dachineni R, et al. In Vitro and In Vivo Antimetastatic Effect of Glutathione Disulfide Liposomes. Cancer Growth Metastasis 2017;10:1179064417695255 [PubMed]
- Wang Y, Wang T, Sun Y, et al. Astrocyte elevated gene-1 promotes tumour growth and invasion by inducing EMT in oral squamous cell carcinoma. Sci Rep 2017;7:15447. [Crossref] [PubMed]
- Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 2009;7:147-54. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Zhu S, Wu H, Wu F, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008;18:350-9. [Crossref] [PubMed]
- Tavazoie SF, Alarcón C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147-52. [Crossref] [PubMed]
- Lin LL, Wang W, Hu Z, et al. Negative feedback of miR-29 family TET1 involves in hepatocellular cancer. Med Oncol 2014;31:291. [Crossref] [PubMed]
- Li G, Zhang W, Gong L, et al. MicroRNA-125a-5p Inhibits Cell Proliferation and Induces Apoptosis in Hepatitis B Virus-Related Hepatocellular Carcinoma by Downregulation of ErbB3. Oncol Res 2017; [Epub ahead of print]. [Crossref] [PubMed]
- El Bezawy R, Cominetti D, Fenderico N, et al. miR-875-5p counteracts epithelial-to-mesenchymal transition and enhances radiation response in prostate cancer through repression of the EGFR-ZEB1 axis. Cancer Lett 2017;395:53-62. [Crossref] [PubMed]
- Zhang T, Cai X, Li Q, et al. Hsa-miR-875-5p exerts tumor suppressor function through down-regulation of EGFR in colorectal carcinoma (CRC). Oncotarget 2016;7:42225-40. [PubMed]
- He XX, Chang Y, Meng FY, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012;31:3357-69. [Crossref] [PubMed]
- Santamaría E, Muñoz J, Fernández-Irigoyen J, et al. Toward the discovery of new biomarkers of hepatocellular carcinoma by proteomics. Liver Int 2007;27:163-73. [Crossref] [PubMed]
- Michon F, Tummers M, Kyyrönen M, et al. Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev Biol 2010;340:355-68. [Crossref] [PubMed]
- Jia XQ, Cheng HQ, Qian X, et al. Lentivirus-mediated overexpression of microRNA-199a inhibits cell proliferation of human hepatocellular carcinoma. Cell Biochem Biophys 2012;62:237-44. [Crossref] [PubMed]
- Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem 2009;284:32015-27. [Crossref] [PubMed]
- Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008;27:5651-61. [Crossref] [PubMed]
- Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A 2010;107:264-9. [Crossref] [PubMed]
- Emdad L, Sarkar D, Su ZZ, et al. Astrocyte elevated gene-1: recent insights into a novel gene involved in tumor progression, metastasis and neurodegeneration. Pharmacol Ther 2007;114:155-70. [Crossref] [PubMed]
- Emdad L, Lee SG, Su ZZ, et al. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci U S A 2009;106:21300-5. [Crossref] [PubMed]
- Callegari E, Elamin BK, Sabbioni S, et al. Role of microRNAs in hepatocellular carcinoma: a clinical perspective. Onco Targets Ther 2013;6:1167-78. [PubMed]


