
Distribution of HLA-A alleles and its relation to clinical outcome in Uyghur and Han patients with advanced squamous cell cervical cancer in Xinjiang, China
Introduction
Cervical cancer (CaCx) is the third most common malignancy among women worldwide (1). From 1930 to 2011, the mortality of CaCx has been continuously decreasing as a result of preventive screening and early detection of cancer (2). However, in China, there were approximately 98,900 newly diagnosed CaCx cases and 30,500 deaths due to CaCx in 2015 (3). CaCx accounts for about one-third of worldwide morbidity and mortality (4-6). Thus, CaCx remains a threat to women’s health.
The residents of Xinjiang, China, mainly consist of Uyghur and Han populations. Uyghur women displayed the highest morbidity and mortality due to CaCx among Chinese women (7), making CaCx a disease with an especially high incidence in this area, which is partly a result of widespread human papillomavirus (HPV) infection (7). It is known, however, that HPV infection itself is not sufficient to induce tumorigenesis (8), and there are likely additional factors that contribute to the development of CaCx. Human leukocyte antigens (HLAs), a host genetic factor and critical node of the host immune system, play an important role in T cell immunity. The HLA system includes gene loci located on the short arm of chromosome 6 at 6p21.3 (9). HLA-A belongs to the classical HLA-I molecules. They can present HPV-derived peptides to CD8+ T cells, playing a vital role in the suppression of HPV infection and the development of CaCx. A number of studies have been performed to uncover the relationship between HLA-A alleles and various types of disease (10-14), the conclusions of which have been varied for different populations. Several studies have examined the associations between CaCx and HLA in the Xinjiang population, but most of these studies focused on HLA class II molecules (15,16). Thus, knowledge of any correlations between the frequencies of HLA-A alleles and CaCx in Uyghur women is limited. Moreover, there is no report available yet on HLA-A alleles and clinical outcomes in CaCx patients.
Most of the Uyghur females with CaCx were in the middle or late stage of the disease when first admitted to the hospital and were unwilling to be treated. These factors contributed to a disproportionate level of morbidity and mortality among Uyghur CaCx patients. Therefore, it would be beneficial to determine factors that contribute to the onset of CaCx. In this study, we recruited patients with advanced squamous cell cervical cancer (SCC, stage IIb–IVb), as well as control healthy subjects, from both Uyghur and Han populations. The frequencies of HLA-A alleles were determined in the two ethnic populations with or without SCC. HLA-A alleles were also analyzed in relation to HPV status and clinical outcome.
Methods
Subjects
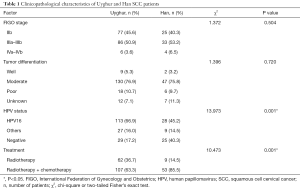
Our study subjects consisted of 231 women (169 Uyghur and 62 Han individuals) who were admitted to the Department of Gynecologic Radiotherapy, Affiliated Tumor Hospital of Xinjiang Medical University for cancer screening and initial treatment between May 2012 and March 2016. The criteria of International Federation of Gynecology and Obstetrics (FIGO) in 2009 (17) were used for tumor staging, and all patients were histologically diagnosed with SCC and FIGO stage in IIb–IVb. The patients received the initial therapy of radiotherapy or concurrent radiochemotherapy at our hospital. SCC patients who had any CaCx treatment history, such as surgery, radiotherapy, chemotherapy or immunotherapy, or were simultaneously diagnosed with other cancers, were excluded (Table 1). A total of 197 healthy women without a family history of CaCx (101 Uyghur and 96 Han individuals) were recruited as controls from physical examination center of the same hospital.
Full table
HPV and HLA typing
All SCC patients were characterized in terms of HPV status and HLA genotypes. Vaginal secretions were taken from patients to determine HPV status. All patients were informed that they should not use vaginal drugs, wash the vagina, or participate in sexual activity 3 days before the secretion was taken. Two gynecologists exposed the cervix of the uterus and took samples using a disposable cervical cell collector (ChaozhouKaipu Biochemistry Co. Ltd., China). The HPV subtype in samples was determined by the Hybrimax HPV DNA detection method.
Genomic DNA was obtained from peripheral whole blood. The DNA was extracted with whole blood genomic DNA extraction kits [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China]. The DNA concentration and purity were determined with a UV spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA with a concentration of 0.5 µg/µL and purity in the range of 1.7–1.9 was considered to be acceptable for HLA-A typing. If the DNA did not reach these requirements, the DNA was re-extracted. The final product was stored at −20 °C before genotyping. HLA-A alleles were genotyped with a HLA sequencing commercial kit by polymerase chain reaction sequence-based typing (PCR-SBT) according to the manufacturer’s instructions (experimental procedures of genotyping HLA alleles were completed in BGI Clinical Laboratories, Shenzhen, China). Briefly, following PCR amplification, the amplified products were purified by enzyme digestion and used as a template for sequencing reactions for exons 2, 3, and 4. Sequencing reaction products were electrophoresed on DNA analyzer and the data were analyzed with sequence analysis software.
Clinical treatment and follow-up
All of the patients in this study received radiotherapy or concurrent radiochemotherapy as the primary treatment for SCC. For radiotherapy, extracorporeal irradiation (intensity modulated radiation therapy 50 Gy/25–28 Fx or 3-dimensional conformal radiation therapy at a dosage of 45–50 Gy, if imaging or pathology showed metastatic lymph nodes in the pelvic cavity, locally added to 55–60 Gy) and intracavitary irradiation (intracavitary after loading therapy with a dose of 36–42 Gy at point A, 6–7 times/week). On the day of intracavitary after loading therapy, extracorporeal irradiation therapy was not given. For chemotherapy, PVB (50 mg/m2 cisplatin +30 mg/m2 bleomycin hydrochloride +1.5 mg/m2 vincristine sulfate) or TP (135 mg/m2 taxol +50 mg/m2 cisplatin) were used. Three cycles of chemotherapy were performed. After completion of therapy, patients were followed at 3-month intervals during the first 2 years and at 6-month intervals thereafter. Survival and recurrence data were collected for all patients. Disease-specific survival (DSS) was defined as the time from diagnosis to death due to SCC. Disease-free survival (DFS) was defined as the time from diagnosis until the date of first disease recurrence (local, regional, distant or a combination) or last follow-up in case of no recurrence or disease specific death.
Statistical analysis
SPSS software (version 21.0, IBM Corporation, Armonk, NY, USA) was used for statistical analyses. Age comparisons were performed with the Student’s t-test. Allele frequencies were calculated by direct counting. Chi-square or two-tailed Fisher’s exact tests were performed to analyze the clinicopathological characteristics of the two ethnic groups and analyze the different distributions of HLA-A alleles. The risk associations were determined by odds ratios (ORs) with 95% confidence intervals (CIs). The Kaplan-Meier method was used to estimate DSS and DFS, and the groups were compared using the log-rank test. Multivariate Cox regression was performed to examine the independent risk factors, including all variables. P<0.05 was considered statistically significant in two-sided tests.
Results
The mean age of the patients at diagnosis was 52.95±10.48 years (range, 31.0–81.0 years) while that of control subjects was 50.50±9.75 years (range, 29.0–71.0 years). In the cancer group, the mean age of the Uyghur patients was 53.97±10.74 years (range, 31.0–81.0 years), and that of the Han patients was 52.57±10.39 years (range, 35.0–75.0 years). There was no statistically significant difference between the two groups in terms of age at SCC onset (t=0.895, P=0.372). The Uyghur patients were significantly different from the Han patients in HPV status and treatment condition while no statistically significant difference was apparent in FIGO stage and tumor differentiation. The Uyghur patients were more frequently infected with HPV16 or other HPV types compared with the Han patients (P=0.001). The Han patients responded better to systematic treatment than the Uyghur patients (P=0.001). The clinicopathological characteristics of the Uyghur and Han patients with advanced SCC are shown in Table 1.
HLA-A distribution
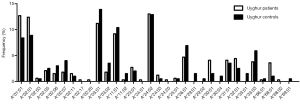
In total, 28 HLA-A alleles were identified in the Uyghur and Han control subjects (Figure 1). The most common HLA-A allele in the Uyghur control subjects was A*03:01 (13.9%) while A*24:02 was the most common allele in the Han control subjects (22.4%). The frequency of A*03:01 in the Uyghur control subjects was significantly higher than the Han control subjects (4.2%, P=0.001). On the other hand, the frequency of A*24:02 in the Han control subjects was significantly higher than the Uyghur control subjects (12.9%, P=0.013). Moreover, the frequency of A*01:01 in the Uyghur control subjects was 8.4%, compared with 3.1% in the Han control subjects. The frequency of A*03:02 in the Uyghur control subjects was 3.5% while it was not detected in the Han control subjects, the differences of which were both statistically significant (P=0.025 and P=0.015). In contrast, the frequencies of the A*11:01 and A*30:01 alleles in the Han control subjects were higher than the Uyghur control subjects (18.2% vs. 10.4%, P=0.026 and 11.5% vs. 1.5%, P<0.001, respectively). These results indicated that ethnicity played a role in the distribution of the HLA-A alleles.
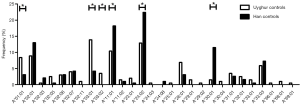
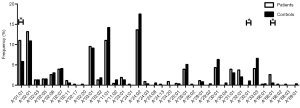
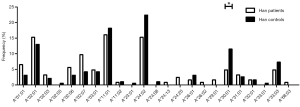
Different HLA-A allele frequencies also existed between SCC patients and control subjects. A total of 36 alleles were genotyped in the patients or control subjects. HLA-A*24:02 was the most common allele in both SCC patients and control subjects (13.6% vs. 17.5%). However, no statistically significant difference was found in the frequency of this allele between the two groups (P=0.118). Different frequencies were evident between SCC patients and control subjects for some HLA-A alleles. It is worth noting that the alleles A*01:01 and A*68:01 were more frequently seen in the patients than the control subjects, suggesting that they might be considered as risk factors for SCC (11.0% vs. 5.8%, P=0.007, OR =2.002, 95% CI: 1.200–3.340 for A*01:01 and 2.6% vs. 0.5%, P=0.033, OR =5.227, 95% CI: 1.163–23.496 for A*68:01). In addition, the frequency of A*33:01 in the control subjects was 1.0%, whereas it was not detected in SCC patients. A significant association between A*33:01 and SCC was found when comparing the two groups (0.0% vs. 1.0%, P=0.045) (Figure 2). When subjects with or without SCC were compared within their individual ethnic groups, different patterns were evident between the Uyghur and Han women. A total of 32 alleles were detected in the Uyghur subjects, while only 23 alleles were detected in the Han subjects. Compared with the Han SCC patients, the Han control subjects had a significantly higher chance of carrying the allele A*30:01, suggesting a protective role of A*30:01 for SCC (4.8% vs. 11.5%, P=0.043, OR =0.393, 95% CI: 0.155–0.999) (Figure 3). On the contrary, there was no statistically significant difference in the frequencies of any HLA-A alleles between the Uyghur patients and the control subjects (Figure 4). These results suggested that different HLA-A alleles were responsible for conferring susceptibility to, or protection against SCC in different ethnic groups, indicating genetic factors might influence onset of the disease.
HLA-A distribution, HPV infection status, and disease outcome
When analyzing the association between the distribution of HLA-A alleles and HPV status, we did not identify any HLA-A alleles correlated with HPV16 in any of the SCC patients (data not shown). At the end of our follow-up (July 2017), we had lost contact with 40 patients (32 Uyghur and 8 Han women) because their phone number was wrong or had been canceled. Thus, the lost follow-up rate was 17.3% (18.9% for Uyghur and 12.9% for Han patients). One Uyghur patient died of intestinal obstruction during treatment. Finally, follow-up information from 190 patients was obtained. Meanwhile, information on FIGO stage, tumor differentiation and treatment condition were collected. At this point, the median follow-up of the patients was 25.5 months (range, 3.0–61.0 months) (25.0 months for Uyghur and 27.0 months for Han). The results of the Kaplan-Meier curves and log rank tests indicated that the Uyghur and Han SCC patients had similar DSS and DFS (P=0.153 and P=0.219) (data not shown). However, a higher DSS was observed in patients with the A*30:01 allele if the Uyghur and Han patients were analyzed together. More specifically, of the 16 patients carrying this allele, only one died of SCC. Of the 174 patients who did not carry the allele, 40 died of SCC. The DSS of A*30:01 carriers and non-carriers were 93.8% and 77.0%, respectively, with a significant difference between the two groups (P=0.044) (Figure 5A). The presence of A*30:01 also showed a tendency to correlate with increased DFS among all advanced SCC patients. One of the 16 patients with the A*30:01 had a recurrence while 38 of the 174 patients who were without this allele had recurrence. The DFS of A*30:01 carriers and non-carriers were 93.8% and 78.2%, respectively (P=0.060) (Figure 5B). The patients with the A*68:01 allele tended to demonstrate poorer DSS and DFS compared to non-carrier patients. To be specific, of the nine patients carrying this allele, the four died of SCC. Of the 181 patients who did not carry the allele, 37 died of SCC. The DSS of A*68:01 carriers and non-carriers were 55.6% and 79.6%, respectively (P=0.065). Four of the nine patients with A*68:01 allele had a recurrence while 35 of the 181 patients who were without this allele had recurrence. The DFS of A*68:01 carriers and non-carriers were 55.6% and 80.7%, respectively (P=0.079) (Figure 5C,D). No differences were observed with the other alleles when the two ethnic groups were analyzed together. When Uyghur or Han patients were compared with the control subjects from the same ethnic group, Uyghur patients carrying A*30:01 allele displayed a tendency for higher DSS (carriers vs. non-carriers, 100% vs. 73.8%, P=0.069) (Figure 5E), whereas no difference was observed in Han patients or for other alleles. In order to avoid the effect of treatment on patient prognosis, 139 patients treated with concurrent radiochemotherapy and 51 patients treated with radiotherapy were analyzed separately for survival analysis. The presence of A*30:01 showed a tendency to correlate with increased DSS among the patients only received radiotherapy (carriers vs. non-carriers, 100% vs. 57.4%, P=0.079) when there is no statistical difference in patients who received concurrent radiochemotherapy (Figure 5F).
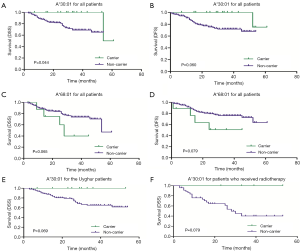
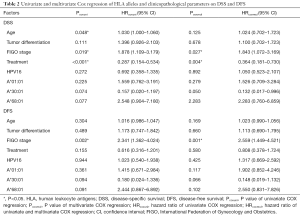
Univariate Cox proportional hazards model analysis was performed to determine prognostic factors for DFS and DSS on all SCC patients. The factors investigated included HLA gene distribution and other already known risk factors, such as age, FIGO stage, tumor differentiation, treatment condition, and HPV status. Treatment condition was significantly related to an increased DSS (P<0.001, HR =0.287, 95% CI: 0.154–0.534) while age and FIGO stage were negatively correlated to DSS (P=0.048, HR =1.030, 95% CI: 1.000–1.060 and P=0.019, HR =1.878, 95% CI: 1.109–3.179, respectively). Advanced FIGO stage was the only factor that was significantly related to a decreased DFS (P=0.002, HR =2.341, 95% CI: 1.362–4.024). No significant difference was observed between the A*30:01 and A*68:01 carriers in terms of DSS and DFS, though there was a tendency for them to relate to DSS and DFS (P=0.074 and P=0.077 for DSS, P=0.094 and P=0.091 for DFS). To determine if these factors were independent prognostic factors for advanced SCC, multivariate Cox regression analysis also was performed on all SCC patients. Results showed that FIGO stage and treatment condition were two independent predictors for DSS (P=0.027, HR =1.843, 95% CI: 1.072–3.169 and P=0.004, HR =0.364, 95% CI: 0.181–0.730, respectively) while only FIGO stage was an independent predictor for DFS (P=0.001, HR =2.559, 95% CI: 1.449–4.521) (Table 2). A*30:01 showed a tendency to be independent protective predictor for DSS (P=0.050, HR =0.132, 95% CI: 0.017–0.996). When the Uyghur and Han SCC patients were analyzed separately, no HLA-A allele was found to be significantly related to DSS and DFS (data not shown).
Full table
Discussion
A number of studies have demonstrated that HLA-A alleles are associated with susceptibility or protection against CaCx. However, various conclusions have been drawn from studies conducted among different populations on the association of HLA-A alleles with CaCx. The diverse genetic composition of study subjects among different studies might be one reason for the inconsistency. With respect to HLA-A alleles, a significantly elevated risk for SCC was observed in American populations with the A*03:01 allele (OR =1.4, 95% CI: 1.1–1.9) (10). With high-resolution analysis, 39 HLA-A alleles were identified in Indian populations and A*02:01, A*02:35, and A*33:01 were found to be associated with an increased risk, while A*24:44 and A*32:01 were associated with a decreased risk for CaCx (11). In a study conducted among Han women, a total of 28 alleles for HLA-A were genotyped, although none of the alleles were statistically correlated with the onset and development of SCC (18). A study performed on Japanese women reported that the allele A*24:02 was associated with an increased risk of CaCx (19). A study in Chinese women from Hong Kong on the association between the distribution of HLA-A alleles and CaCx incidence reported that HLA-A*02:07/A*24:02 was associated with a decreased risk whereas, HLA-A*11:04 was associated with an increased risk for CaCx (20). Significant differences of in HLA-A allele frequencies between CaCx patients and healthy females in Switzerland have also been reported (21). The results of many of these studies are inconsistent, however, especially with respect to different countries and different races. In some cases, the same race in different regions has produced different results. This may be different from the genetic diversity of women in different countries, regions, and races. It may also be related to the inclusion and exclusion criteria, the selection criteria of the control group, the number of cases, and the testing methods used in these studies. These reasons lead to deviations and even conflicting results. Although some conflicts are apparent in the results, it is still apparent that HLA-A alleles are correlated with the incidence of CaCx.
The incidence of CaCx among the Uyghur population is 527/100,000, which is much higher than the average incidence of 126.94/100,000 in China as a whole country. Positive and negative correlations of HLA genes with CaCx have been reported among the Uyghur population (15). However, HLA-A alleles have not been studied as extensively in Uyghur women from China. In this study, we found that different HLA-A allele distributions exist between Uyghur and Han women control subjects, suggesting distinct HLA allele distributions exist in different ethnic groups. These distinct alleles may contribute to the unusual high incidence of SCC among Uyghur women.
In this study, we found that a different HLA-A distribution existed between the control subjects from the Uyghur and Han populations. Thus, we propose that ethnicity plays a role in the distribution of HLA-A alleles. The comparison between 231 SCC patients and 197 control subjects from the Uyghur and Han populations led to the finding that the alleles A*01:01 and A*68:01 were more frequently observed in SCC patients, suggesting they might be responsible for an increased susceptibility to advanced SCC (P=0.007, OR =2.002 and P=0.033, OR =5.227). However, no statistically significant difference was found when women with or without SCC were compared within their own ethnic groups (Uyghur patients and control subjects: 12.7% vs. 8.4%, P=0.123 for A*01:01, 3.6% vs. 1.0%, P=0.126 for A*68:01. Han patients and control subjects: 6.5% vs. 3.1%, P=0.161 for A*01:01 while no distribution of A*68:01 was observed). This might be caused by the relatively small sample cohort size in populations with complex genetic backgrounds living in Xinjiang, China. Interestingly, the frequency of A*01:01 in the Uyghur control subjects was higher compared with Han control subjects (8.4% vs. 3.1%, P=0.025). We also found this allele was enriched in Uyghur patients compared with the Uyghur control subjects. This might partly explain why the Uyghur women are more susceptible to CaCx than Han women. It will be worthwhile elucidate the mechanism behind the connection between A*01:01 and CaCx in future studies.
The allele A*33:01 was only identified in control subjects (1.0%), making its frequency significantly different from that in SCC patients (P=0.045), suggesting this allele might serve as a protective gene for advanced SCC. It is worth noting that A*33:01 was more common in Uyghur control subjects than Uyghur SCC patients, although the difference was not statistically significant (P=0.052). The difference may become more significant with a larger sample size. It will also be worthwhile to test if this allele protects against advanced SCC. Moreover, we noticed that the frequency of A*30:01 in Han patients was significant lower than in Han control subjects (4.8% vs. 11.5%, P=0.043, OR =0.393), which suggested that A*30:01 was a protective allele for Han women. However, no similar pattern for A*30:01 was observed between Uyghur patients and control subjects (4.1% vs. 1.5%, P=0.145). It is also worth mentioning that the frequency of the A*30:01 allele in Uyghur control subjects was significantly lower compared with Han control subjects (1.5% vs. 11.5%, P<0.001). The decreased chance of having A*30:01 allele in the Uyghur women may have contributed to the relatively high incidence of SCC. There are approximately 15 high-risk HPV types that cause approximately 95% of all cervical carcinomas. HPV16 accounts for about 50% of all cases (22). In this study, however, there was no statistically significant association between HLA-A alleles and HPV16 status.
In addition, we evaluated the relationship between HLA-A alleles and clinical outcome in the Uyghur and Han patients with advanced SCC. Results of univariate and multivariate Cox regression indicated that several clinicopathological parameters were related to DSS or DFS in patients regardless of the ethnicity (Table 2). None of the HLA-A allele was found to be significantly related to DSS or DFS. A*30:01 was marginally related to onset and development of advanced SCC. We believe that with a larger sample size, we might be able to obtain a clearer picture of the relationship between A*30:01 and SCC. In summary, this is the first report of the differences in the distribution of HLA-A alleles in the Uyghur and Han women with advanced SCC living in Xinjiang, China. Our results suggest that in women from the Uyghur and Han populations, different HLA-A alleles may contribute to the development of advanced SCC and to some extent may influence the prognosis. These results confirm the complexity and genetic polymorphisms of the HLA system, and further confirm that different frequencies of expression of HLA alleles associated with SCC exists between two ethnic groups with different genetic backgrounds, such as Uygur and Han women. Some HLA-A alleles were not statistically significantly associated with SCC in this study, but a tendency was discernible. In future research, expanding the cohort size will help to test the hypothesis generated in this study.
Conclusions
Our findings indicated that two ethnic populations living in the same geographic area displayed different characteristics of the same cancer. In addition, individuals from two ethnic groups displayed different HLA-A gene distributions. HLA-A alleles may affect the occurrence and development of advanced SCC in both the Uyghur and Han women. More specifically, the alleles A*01:01 and A*68:01 are potential susceptibility factors for advanced SCC while HLA-A*33:01 might serve as a protective allele. The allele A*30:01 might be an independent predictor of advanced SCC.
Acknowledgments
Funding: This work was supported by the Regional Joint Fund of the National Natural Science Foundation of China (No. U1603282) and Ministry of Science and Technology International Cooperation Project (No. 2012DFA31560).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.27). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study protocol for the collection of samples and clinical information was approved by the Institutional Ethics Committee of the Affiliated Tumor Hospital Xinjiang Medical University (No. G-201603 and B-20101201). Written informed consent was obtained from each of participants before enrollment in the study to authorize the collection of blood and its use as source of DNA for HLA typing.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang Q, Qin J, Chen A, et al. Downregulation of microRNA-145 is associated with aggressive progression and poor prognosis in human cervical cancer. Tumour Biol 2015;36:3703-8. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer Statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Xu QX, Zhang ZY. High-risk human papillomavirus genotypes in cervical lesions and vaccination challenges inChina. Asian Pac J Cancer Prev 2015;16:2193-7. [Crossref] [PubMed]
- Xu F, Cao M, Shi Q, et al. Integration of the full-length HPV16 genome in cervical cancer and Caski and Siha cell lines and the possible ways of HPV integration. Virus Genes 2015;50:210-20. [Crossref] [PubMed]
- Chen W, Zheng R, Zeng H, et al. Annual report on status of cancer in China, 2011. Chin J Cancer Res 2015;27:2-12. [Crossref] [PubMed]
- Haimiti A, Hailiman Y, Gulina A, et al. Reduced expression of members of the mhc-i antigen processing machinery in ethnic Uighur women with cervical cancer in the Xinjiang region of China. Curr Oncol 2014;21:e67-74. [Crossref] [PubMed]
- Gimenes F, Teixeira JJ, de Abreu AL, et al. Human leukocyte antigen (HLA)-G and cervical cancer immunoediting: a candidate molecule for therapeutic intervention and prognostic biomarker? Biochim Biophys Acta 2014;1846:576-89. [PubMed]
- Wang RZ, Zhang DG, Wu R, et al. HLA-A*02-B*46 haplotype: an adverse prognostic factor in Han patients with nasopharyngealcarcinoma. J Huazhong Univ Sci Technolog Med Sci 2016;36:700-4. [Crossref] [PubMed]
- Madeleine MM, Johnson LG, Smith AG, et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res 2008;68:3532-9. [Crossref] [PubMed]
- Gokhale P, Mania-Pramanik J, Sonawani A, et al. Cervical cancer in Indian women reveals contrasting association among common sub-family of HLA class I alleles. Immunogenetics 2014;66:683-91. [Crossref] [PubMed]
- Näsman A, Andersson E, Marklund L, et al. HLA Class I and II expression in oropharyngeal squamous cell carcinoma in relation to tumor HPV status and clinical outcome. PLoS One 2013;8:e77025 [Crossref] [PubMed]
- Lyke KE, Fernández-Viňa MA, Cao K, et al. Association of HLA alleles with Plasmodium falciparum severity in Malian children. Tissue Antigens 2011;77:562-71. [Crossref] [PubMed]
- Kang EH, Kim JY, Takeuchi F, et al. Associations between the HLA-A polymorphism and the clinical manifestations of Behcet’s disease. Arthritis Res Ther 2011;13:R49. [Crossref] [PubMed]
- Hu JM, Sun Q, Li L, et al. Human leukocyte antigen-DRB1*1501 and DQB1*0602 alleles are cervical cancer protective factors among Uighur and Han people in Xinjiang, China. Int J Clin Exp Pathol 2014;7:6165-71. [PubMed]
- Guzalinuer A, Mihrinsa A, Zhang SQ, et al. Association between HPV infection and HLA-DQB1 alleles polymorphism in the cervical carcinogenesis in Uyghur women in southern Xinjiang. Zhonghua Zhong liu Za Zhi 2010;32:492-6. [PubMed]
- Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet 2009;105:103-4. Erratum in: Int J Gynaecol Obstet 2010;108:176. [Crossref] [PubMed]
- Xiao X, Liu L, Li WJ, et al. HLA-A, HLA-B, HLA-DRB1 polymorphisms and risk of cervical squamous epithelial cellcarcinoma: a population study in china. Asian Pac J Cancer Prev 2013;14:4427-33. [Crossref] [PubMed]
- Hosono S, Kawase T, Matsuo K, et al. HLA-A alleles and the risk of cervical squamous cell carcinoma in Japanese women. J Epidemiol 2010;20:295-301. [Crossref] [PubMed]
- Chan DP, Cheung TH, Tam AO, et al. Risk association between human leukocyte antigen-A allele and high-risk human papillomavirus infection for cervical neoplasia in Chinese women. J Infect Dis 2005;192:1749-56. [Crossref] [PubMed]
- Ghaderi M, Nikitina Zake L, Wallin K, et al. Tumor necrosis factor A and MHC class I chain related gene A (MIC-A) polymorphisms in Swedish patients with cervical cancer. Hum Immunol 2001;62:1153-8. [Crossref] [PubMed]
- Rosales R, Rosales C. Immune therapy for human papillomaviruses-related cancers. World J Clin Oncol 2014;5:1002-19. [Crossref] [PubMed]


