Genomic alterations across six hepatocellular carcinoma cell lines by panel-based sequencing
Introduction
Hepatocellular carcinoma (HCC), the most common type of liver cancer, is the third leading cause of cancer-related death globally (1). Hepatocarcinogenesis is a complex multistep process driven by the accumulation of genetic alterations in oncogenic genes and pathways (2,3). In recent years, multicenter and multi-omic cancer genome projects based on next-generation sequencing (NGS) have exposed the comprehensive mutational landscape and core oncogenic network of HCC patients, including aberrant signaling cascades, such as the p53 cell cycle, Wnt/β-catenin and MAPK pathways (4,5). A number of cell lines established from HCC tumors have been extensively used as experimental models for this disease, with the advantage of a relative homogeneity of the genetic traits within a cell colony compared to tissues. However, little attention has focused on the mutational patterns in HCC cell lines.
Conventional methods, such as whole-genome sequencing, provide extensive variation landscapes of the cancer genome; however, these results are obtained at the expense of relatively low coverage and a large amount of redundancy data, which are obstacles to the application of these methods in clinical practice (6). Gene panel testing is an emerging option for searching for genetic variants of specific genes of interest (7-9). Gene panel testing has several remarkable advantages compared to full-genome sequencing, including deeper sequencing, higher coverage for rare variant identification and the generation of more manageable data (6).
Therefore, we established a panel-based sequencing platform targeting 325 genes mutated in various types of cancer to identify somatic mutations in six HCC cell lines. This study aimed to test the performance of the targeted sequencing platform for identification of genetic variants in cancer-related genes and, more importantly, to explore the somatic variations in HCC experimental cellular models compared to previous knowledge of mutation status in HCC tissues.
Methods
Cell line culture
A total of six human hepatoma cell lines were used. The HepG2 cell line was originally obtained from the American Type Culture Collection (Manassas, VA, USA). HuH-7, Hep3B and SK-HEP-1 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The MHCC97 series cells (MHCC97H and MHCC97L) cell lines were kindly provided by the Institutes of Biomedical Sciences Fudan University (Shanghai, China). The HepG2.2.15 cell line was kindly provided by Prof. J. T. Guo, Drexel Institute for Biotechnology and Virology Research, Drexel University College of Medicine (PA, USA). The complete growth medium for the cells was supplemented with 10% fetal bovine serum and penicillin-streptomycin antibiotics (Gibco, Grand Island, NY, USA). The cells were incubated in a humidified atmosphere containing 5% CO2 at 37 °C.
Targeted NGS Panel design
Genes included in our panel were selected according to the following criteria: cancer-related genes involved in core signaling processes and pathways collected from scientific literature and genes with high somatic mutation frequency from the Catalogue of Somatic Mutations in Cancer database (COSMIC, http://www.sanger.ac.uk/cosmic) (10). With these criteria, the panel consisted of the protein coding regions of 325 genes (Available online: http://tcr.amegroups.com/public/addition/tcr/supp-tcr.2018.02.14-1.pdf) with a total length of 901 kb of target regions, using the hg19/GRCh37 genome build (Ensembl version 75). A custom capture reagent containing predefined probe sets was constructed using the SeqCap EZ Choice Library according to the NimbleDesign guidelines (Roche NimbleGen, Madison, WI, USA).
Target enrichment of genomic DNA and sequencing
Genomic DNA was extracted from the cell lines using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The DNA sample library was prepared using a KAPA Library Preparation Kit (KAPA Biosystem, Woburn, MA, USA) according to the manufacturer’s guidelines. Each sample was fragmented to 180–220 bp, followed by end-repair, A-tailing and Illumina adapter ligation (Life Technologies, Carlsbad, CA, USA). After dual-SPRI size selection (250–450 bp), the libraries were amplified by polymerase chain reaction (PCR) and quantified using Qubit 3.0 (Life Technologies) and an Agilent 2100 Bioanalyzer DNA 1000 Kit (Agilent Technology, California, USA). The amplified sample library was used to enrich target regions by hybrid capture for the 325 gene target regions, using a solution-based SeqCap EZ Choice Library Kit (Roche NimbleGen). Finally, the amplified and captured multiplex DNA samples were subjected to paired-end 150-bp read length sequencing using a HiSeq X Ten platform (Illumina, San Diego, CA, USA) following the manufacturer’s instructions.
Bioinformatics analysis
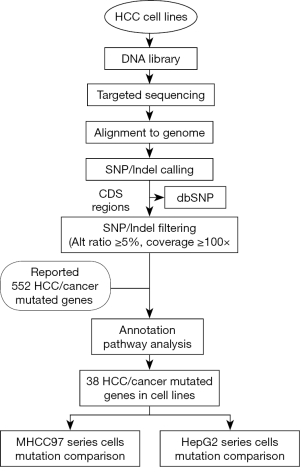
The bioinformatics analysis flowchart was developed as follows (Figure S1). The captured sequencing data were mapped against the hg19 human reference genome using the Burrows-Wheeler Aligner (BWA) (11). SAMtools was used to generate pileup files and VarScan2 was used to call mutations, including single nucleotide variants (SNV) and short insertions/deletions (indels) (12). To identify somatic mutations in the exome region, only variants located in the coding DNA sequence (CDS) regions were used for further analysis. We filtered out known polymorphisms documented in dbSNP129. To minimize the potential errors in base calling, we defined the following filtering conditions: (I) variants of the candidate position with an alteration frequency no less than 5% and (II) sites with at least a 100 read depth.
Pathway analysis
We performed both computation-based and literature-based pathway analyses for annotating the candidate mutated genes in HCC cell lines. We used Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) for BBID, BioCarta and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping (13). Well-studied pathways, including the p53 cell cycle, Wnt/β-catenin, chromatin remodeling, MAPK and Notch, were surveyed by referring to prior knowledge and literature.
Results
Cell line characteristics
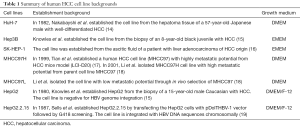
The establishment background of the HCC cell lines is summarized (14-19) (Table 1), including three cell lines derived from biopsies of HCC patients (HuH7, Hep3B and HepG2), one derived from ascitic fluid of liver adenocarcinoma patient (SK-HEP-1), one from HepG2 cells (HepG2.2.15) and MHCC97 series cells (MHCC97L and MHCC97H). MHCC97L and MHCC97H had similar genetic backgrounds but different metastatic potentials as a result of in vivo selection of MHCC97, which provided metastatic human HCC models (20). The cell line HepG2.2.15 is a widely used hepatitis B viral (HBV) replication model that was established by stably transfecting an HBV genome into HepG2 cells (19).
Full table
Sequencing profile and overall mutations
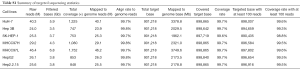
On average, each library yielded 4.5 Gb of sequence, and the mean read depth was approximately 1,000. The average coverage rate in target regions was 99.7%, and the targeted regions covered by at least 100 reads accounted for 99.4% (Table S1).
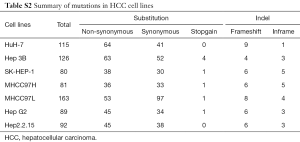
By using bioinformatics analysis and filtering from the raw data, we yielded 100 genes mutated in at least one cell line with an alteration frequency of no less than 5%. A total of 344 non-synonymous mutations, 325 silent mutations, 8 stop-codon-gain mutations, and 69 indels were identified, corresponding to 49 non-silent mutations per cell line (Available online: http://tcr.amegroups.com/public/addition/tcr/supp-tcr.2018.02.14-2.pdf; Table S2). To further identify significantly altered genes in HCC cell lines, we performed a literature-based summary of cancer-related genes from ten high-quality papers published in the last 5 years (4,5,21-28). These research articles and reviews represent comprehensive genomic profiling of HCC and common forms of other human cancers. The aberrant genes that were reported at least once in the above papers were included. A total of 552 HCC and cancer mutated genes were summarized (Available online: http://tcr.amegroups.com/public/addition/tcr/supp-tcr.2018.02.14-3.pdf) and further compared to the 100 mutated genes we identified. We annotated the overlapping 52 genes using both DAVID functional annotation tools (Available online: http://tcr.amegroups.com/public/addition/tcr/supp-tcr.2018.02.14-4.pdf) and eleven major HCC major signaling pathways from the literature (Available online: http://tcr.amegroups.com/public/addition/tcr/supp-tcr.2018.02.14-5.pdf) (4,5,22). A total of 38 candidate genes were identified to be involved in cancer core cellular processes and signaling pathways, such as cell survival, cycle and apoptosis (Table S3).
Mutated cancer-related genes and pathways in HCC cell lines
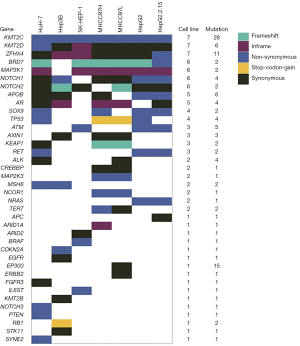
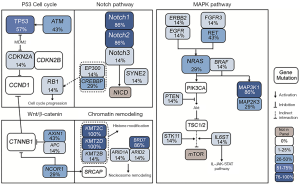
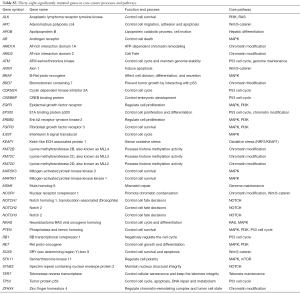
We investigated the 38 genes mutational patterns in the HCC cell lines (Figure 1). Half of the altered genes (27/54) in 11 major HCC signaling pathways (4,5), which are also included in the gene panel, were identified as sequence variations in HCC cell lines, though with different mutational rates to some extent. The mutated genes were significantly enriched in four major HCC signaling pathways (chromatin remodeling, Wnt/β-catenin, MAPK and p53 cell cycle) and one cancer-related pathway (Notch) (Available online: http://tcr.amegroups.com/public/addition/tcr/supp-tcr.2018.02.14-5.pdf; Figure 2). The four major HCC signaling pathways are well-acknowledged as having a high somatic alteration frequency. We identified six significantly mutated genes in the tumor-suppressor p53 cell cycle cascade (TP53, ATM, CREBBP, EP300, CDKN2A and RB1), two genes in the Wnt/β-catenin pathway (AX1N1 and APC) and 11 genes in the MAPK pathway (MAP3K1, MAP2K3, NRAS, RET, FGFR3, ERBB2, EGFR, PTEN, STK11, IL6ST and BRAF). It is worth mentioning that, compared to HCC tissues, more genetic alterations were identified in chromatin modulators (KMT2C, KMT2D, ZFHX4, BRD7 and NCOR1) and the Notch pathway (Notch1 and Notch2). Our results identified three new cancer-related genes that were not significantly mutated in HCC cohorts. These three genes included two tumor suppressors MSH6 (c.2582A>T and c.3005G>A) and SOX9 (c.724A>C and c.715A>C), and one oncogene ALK (c.3600C>G, c.678G>T, c.831G>A and c.931G>C). In short, we have identified the genetic alterations in cancer-related genes and pathways in HCC cell lines by referring to HCC cohorts in previous reports.
Mutational patterns in cell lines from the same parent cells
It was obvious that the four specimen-derived HCC cells from different individuals, including HuH-7, Hep3B, SK-HEP-1 and HepG2, showed different mutational patterns. In contrast, the MHCC97 and HepG2 series of cells showed similar mutational patterns (Figure 1). Therefore, we further analyzed the mutations in these two series of cell lines.
There were 44 mutated genes identified in the MHCC97 series cells (MHCC97H and MHCC97L), and 68% of these genes (30 genes) were found in both cell lines (Figure 3A). We defined genes between which the mutation ratio difference value was less than 5% as “shared mutated genes”. Among the above 38 cancer-related genes, the shared mutated genes between MHCC97L and MHCC97H cells were enriched in oxidative stress (KEAP1), chromatin modification (NCOR1, KMT2D and ARID1A), the Wnt/β-catenin pathway (AXIN1, SOX9) and the MAPK (MAP3K1 and MAP2K3) pathway. However, the genes had several individual somatic variations. ALK and ERBB2 were only mutated in MHCC97L cells, and APOB was only mutated in MHCC97H cells. MHCC97L cells had higher mutation frequency in Notch pathway genes (Notch1 and Notch2) than MHCC97H cells. TERT had higher mutation frequency in MHCC97H cells compared to MHCC97L cells (Figure 3B).
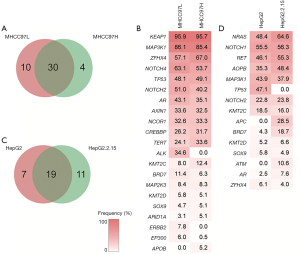
HepG2 and HepG2.2.15 cells showed similar genetic alteration patterns with MHCC97 cells, and 37 mutated genes were detected, with 51% (19 genes) shared among the cell lines (Figure 3C). Two Notch pathway genes (Notch1 and Notch2) and two chromatin regulators (KMT2D and ZFHX4) were identified to have similar alteration frequencies at the same mutation site. Among the 38 genes, there were several individual somatic variations. TP53 was specifically mutated in HepG2 cells, while APC and ATM were specifically mutated in HepG2.2.15 cells (Figure 3D).
Discussion
In our present study, to achieve deep sequencing for rare variant identification in cancer-related genes, we performed a panel-based targeted sequencing method to identify somatic mutations in six widely used HCC cell lines. Our method exhibited a 99.7% mean coverage rate in target regions at an approximately 1,000× read depth, which demonstrated the reliable quality of our custom targeted gene sequencing. We have established a targeted sequencing platform using Roche NimbleGen capture paired with Illumina Hiseq sequencing technology, allowing identification of genetic variants in cancer-related genes for HCC genomic samples.
By comparing the mutated genes in eleven major HCC signaling pathways, we identified a consistency in somatic mutations between HCC tissues and cell lines. To further reveal the critical genomic variations, we focused on 38 significantly mutated genes, selected by pathway enrichment and prior knowledge, that are enriched in five oncogenic pathways. Genetic mutations in these critical genes and pathways may be the molecular mechanisms underlying their carcinogenic characteristics in HCC cell lines. A large proportion of the frequently mutated genes encode multiple chromatin modulators. KMT2C, KMT2D and ARID1A are all potential driver genes in HCC cell lines that are involved in the nucleosome structure remodeling and histone modification of their target genes (5,29). Mutated genes BRD7, ARID1A and ARID2 are core subunits making up the SWI/SNF nucleosome-remodeling complex (30). Aberrant activation of the Notch pathway leads to oncogenesis by promoting the self-renewal of liver cancer stem cells and modification of the inflammatory environment, which may be the cause of HCC (31,32).
In addition to the carcinogenic pathways, we identified three new cancer-related genes that are frequently mutated in HCC cell lines (MSH6, SOX9 and ALK). Deleterious mutations of the mismatch-repair gene MSH6 leads to mismatch repair (MMR) deficiency and microsatellite instability (MSI). MMR deficiency has recently been proved to be a biomarker for predicting clinical benefit of immune checkpoint programmed death 1 (PD-1) blockade for multiple solid tumors, including liver cancer (33,34). It was reported that HCCs have MSI-high (MSI-H) frequency (16%) and MSI-low (MSI-L) frequency (27%), and MSI status might affect HCC patients’ responses to immune checkpoint inhibitors (35,36). Thus, the consequences of MSH6 substitutions in HuH7 and Hep3B (p.Glu861Val and p.Arg1002Lys) remain to be examined further. SOX9 regulates the expression of stemness genes in tumor initiation and invasion in a Wnt/β-catenin-dependent manner (37). ALK mutation is one of the major oncogenic drivers and therapeutic targets in lung cancer, which may also have a potential impact on HCC (38,39). In brief, we identified genetic alterations in HCC cell lines similar to those previously reported in HCC tissues, and we also found several genes and pathways not previously known to be frequently mutated in HCC.
We further observed the different mutational patterns across the four specimen-derived HCC cells from different individuals, Hep3B, HuH-7, SK-HEP-1 and HepG2. These patterns validate that genetic variations exist among different individual tumors of the same tumor type from the perspective of eliminating the distractions associated with tissue complexity (40). In contrast, major similarities and some minor discrepancies in genomic alterations were observed among cell lines from the same clone origin. By comparing the genomic alterations between MHCC97L and MHCC97H, cell strains from the same clone origin but with different metastatic potentials, we found that 68% of shared mutated genes were significantly enriched in core cancer signaling pathways and also identified several differentially mutated genes. Although no specific gene mutations were identified to directly explain their different metastatic potentials, we here verified this type of tumor evolution in cell lines, which is consistent with previous knowledge that HCC primary tumors and metastatic subclones evolve from the same origin in patients (41,42). The two HepG2 series cells had 51% shared mutated genes and several individual somatic variations. The mutational differences between cell lines from the same clone origin may mainly be ascribed to subclones that have been selectively mutated due to their metastatic characteristics or continuous viral replication. Additionally, dynamic mutation accumulation may occur in the long-term selection of in-vitro culture due to genomic instability of tumor cells. In summary, our results validate the theory prevalent in recent years that extensive genetic heterogeneity exists within individual tumors from the perspective of cell lines and that HCC cells from the same clone origin have great similarities in genomic alterations, which may be used to trace HCC cell clone origins.
The genetic variation pattern of HCC cell lines provides insight into these extensively used experimental models. However, HCC cell lines of other clone origins and the detailed molecular mechanism for their mutational similarities and differences need to be further explored to validate the mutation patterns in our study. In future studies, we will further optimize our targeted sequencing platform and explore its application for the research of tumor heterogeneity and clonal evolutionary processes in clinical setting. In summary, we identified altered genes and pathways in HCC cell lines through targeted sequencing technology, which could have implications for the genetic heterogeneity of liver cancer and for tracing tumor origins based on genetic background.
Full table
Full table
Full table
Acknowledgments
We thank Dr Linlin Yan, Minghui Ge, Xiangru Li, Xiaoxi Pan and Xinyuan Zhang at Genecast (Beijing) Biotechnology Co. for assisting bioinformatics analysis and for valuable discussion.
Funding: This work was supported by the National Natural Science Foundation of China (81001072 to H Zhang), the Beijing Natural Science Foundation (7152151 to H Zhang), and the Jiangsu Key Laboratory of Medicine Science and Laboratory Medicine (JSKLM-2014-008 to H Zhang).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.02.14). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Byam J, Renz J, Millis JM. Liver transplantation for hepatocellular carcinoma. Hepatobiliary Surg Nutr 2013;2:22-30. [PubMed]
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. [Crossref] [PubMed]
- Marquardt JU, Thorgeirsson SS. SnapShot: Hepatocellular carcinoma. Cancer Cell 2014;25:550.e1 [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267-73. [Crossref] [PubMed]
- Han SW, Kim HP, Shin JY, et al. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS One 2013;8:e64271 [Crossref] [PubMed]
- Glöckle N, Kohl S, Mohr J, et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur J Hum Genet 2014;22:99-104. [Crossref] [PubMed]
- Easton DF, Pharoah PD, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015;372:2243-57. [Crossref] [PubMed]
- Nikiforova MN, Wald AI, Roy S, et al. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab 2013;98:E1852-60. [Crossref] [PubMed]
- Shepherd R, Forbes SA, Beare D, et al. Data mining using the Catalogue of Somatic Mutations in Cancer BioMart. Database (Oxford) 2011;2011:bar018 [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Nakabayashi H, Taketa K, Miyano K, et al. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 1982;42:3858-63. [PubMed]
- Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980;209:497-99. [Crossref] [PubMed]
- Heffelfinger SC, Hawkins HH, Barrish J, et al. SK HEP-1: a human cell line of endothelial origin. In Vitro Cell Dev Biol 1992;28A:136-42. [Crossref] [PubMed]
- Tian J, Tang ZY, Ye SL, et al. New human hepatocellular carcinoma (HCC) cell line with highly metastatic potential (MHCC97) and its expressions of the factors associated with metastasis. Br J Cancer 1999;81:814-21. [Crossref] [PubMed]
- Li Y, Tang ZY, Ye SL, et al. Establishment of cell clones with different metastatic potential from the metastatic hepatocellular carcinoma cell line MHCC97. World J Gastroenterol 2001;7:630-6. [Crossref] [PubMed]
- Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 1987;84:1005-9. [Crossref] [PubMed]
- Tang ZY, Ye SL, Liu YK, et al. A decade's studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol 2004;130:187-96. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Cleary SP, Jeck WR, Zhao X, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693-702. [Crossref] [PubMed]
- Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet 2012;44:1117-21. [Crossref] [PubMed]
- Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol 2014;11:340-9. [Crossref] [PubMed]
- Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422-33. [Crossref] [PubMed]
- Nault JC, Zucman-Rossi J. Genetics of hepatocellular carcinoma: the next generation. J Hepatol 2014;60:224-6. [Crossref] [PubMed]
- Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8. [Crossref] [PubMed]
- Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760-4. [Crossref] [PubMed]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene 2009;28:1653-68. [Crossref] [PubMed]
- Wang R, Sun Q, Wang P, et al. Notch and Wnt/beta-catenin signaling pathway play important roles in activating liver cancer stem cells. Oncotarget 2016;7:5754-68. [PubMed]
- Lu J, Xia Y, Chen K, et al. Oncogenic role of the Notch pathway in primary liver cancer. Oncol Lett 2016;12:3-10. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Chiappini F, Gross-Goupil M, Saffroy R, et al. Microsatellite instability mutator phenotype in hepatocellular carcinoma in non-alcoholic and non-virally infected normal livers. Carcinogenesis 2004;25:541-7. [Crossref] [PubMed]
- Naboush A, Roman CA, Shapira I, et al. Immune checkpoint inhibitors in malignancies with mismatch repair deficiency: a review of the state of the current knowledge. J Investig Med 2017;65:754-8. [Crossref] [PubMed]
- Larsimont JC, Youssef KK, Sánchez-Danés A, et al. Sox9 Controls Self-Renewal of Oncogene Targeted Cells and Links Tumor Initiation and Invasion. Cell Stem Cell 2015;17:60-73. [Crossref] [PubMed]
- Tan WL, Jain A, Takano A, et al. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol 2016;17:e347-62. [Crossref] [PubMed]
- Jia SW, Fu S, Wang F, et al. ALK gene copy number gain and its clinical significance in hepatocellular carcinoma. World J Gastroenterol 2014;20:183-92. [Crossref] [PubMed]
- Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338-45. [Crossref] [PubMed]
- Miao R, Luo H, Zhou H, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol 2014;61:840-9. [Crossref] [PubMed]
- Xue R, Li R, Guo H, et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 2016;150:998-1008. [Crossref] [PubMed]