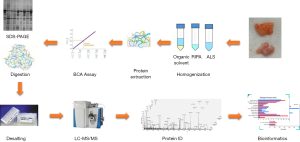
Investigation of an optimal lysis method for the study of thymus and thymoma by mass spectrometry-based proteomics
Introduction
The thymus is a specialized primary lymphoid organ which provides a microenvironment for the development for thymocyte (1). The thymus reaches its maximum weight (20 to 37 grams) by the time of puberty and slowly shrinks with aging, eventually degenerates into tiny islands of adipose tissue. Thymoma, arising from thymic epithelial cells, has been characterized by indolent growth with local invasiveness (2,3). Although the overall incidence of thymoma is relatively low, it is the most common malignancy that develops in the anterior mediastinum and is often associated with autoimmune diseases such as myasthenia gravis (4,5). Thymomas are no longer considered as benign tumors, since they could turn into malignancy indicated by invasive growth, recurrence, and metastases (2,6-8). There is so far no effective treatment for advanced stage or recurrent diseases with poor prognosis (5,9,10).
Several successful studies on thymoma have been reported previously, mainly focusing on various genetic aberrations involved in thymoma (5,11-17). However, protein signaling pathways and networks have not been systematically characterized but are highly demanded towards understanding the development of thymomas. On the technical side, proteomics analysis using mass spectrometry (MS) has been shown as a powerful approach to profile global protein networks in many human tissues and their alterations in many disease states that can be served as clinical biomarkers for diagnosis, prognosis in cancer (18-23). However, large scale proteomics analysis on thymus tissue has not been extensively conducted so far, partially due to difficulties in extracting proteins from the adipose type tissues including thymus. To prepare protein components for proteomics analysis, complex extraction procedures were generally used. In this study, we have developed a simplified and efficient protocol for protein extraction in thymus tissue which facilitated a high coverage of proteome profiles from freshly obtained clinical thymoma tissues using liquid chromatography tandem mass spectrometry (LC-MS) based approaches.
Methods
Tissue sample collection
Frozen thymoma tissue and the para-tumor tissue were obtained from Shanghai Chest Hospital Tissue Biobank. The patient was a 62-year-old male undergoing a VATS tumor resection (tumor size: 6.7×5×3.6 cm) plus thymectomy. Postoperative histological examination revealed a Masaoka-Koga stage IIa WHO type B2 thymoma. The normal thymus tissues were obtained from a 31-year-old male patient undergone cardiovascular surgery. The histotypes of all samples were verified by our pathologists via frozen sections before further processing. All patients have filled out informed consent forms, and all experimental work in this study was approved by the Institutional review board of Shanghai Chest Hospital.
Tissue homogenization and protein extraction
Obtained tissue blocks were cut into very small pieces and 500 µL lysis buffer (corresponding buffer components see Table 1) with protease inhibitor cocktail (Roche, Burgess Hill, UK) was added into each sample. All the sample were stored in lysis buffer overnight at −80 °C for better membrane lysis and then were homogenized in a 2 mL tube with ceramic coated beads using Precellys 24 high-throughput tissue homogenizer (Bertin, FIJI) at 4 °C (highest speed, 30 s shaking, then cool on ice, repeat 3 times). The homogenates were then transferred into precooled 1.5 mL Eppendorf tubes and incubated for 30 minutes on ice. After that, the lysates were spun down at 20,000 g for 30 minutes at 4 °C. All the protein concentrations were determined using the lysate supernatants by standard BCA assay.

Full table
Chloroform/methanol extraction was performed as described before(24), a mixture of 437.5 µL chloroform/methanol (v/v=1:2) was added into sample before homogenization. After that, the samples were kept on ice for 30 minutes, then a mixture of 312 µL H2O/methanol (v/v=1:1) was further added to the mixture. The mixture was then spun at 1,500 g for 5 minutes. The supernatants were transferred and spun down again at 20,000 g for 30 minutes at 4 °C.
For acetone precipitation, 5 volumes of cold acetone were added to the protein sample in a 15 mL falcon tube and vortex thoroughly. The mixture was incubated overnight at −20 °C. After spun down at 15,000 g for 10 minutes, the supernatant was removed and the pellet was carefully washed 3 times with deionized H2O (discard water by pouring) at room temperature and re-solubilized in lysis buffer.
Protein electrophoresis
All the SDS-PAGEs were carried out using a 10% (w/v) gel by the Mini-Protean 3 Tetra Electrophoresis System (Bio-Rad, USA), the gels were stained with silver nitrate and imaged with the ChemiDoc XRS+ system (BIO-Rad, USA).
Protein denature, reduction, alkylation and digestion
6M urea was used to denature the proteins at room temperature for 1 h. Then the proteins were reduced with 5 mM tris (2-carboxyethyl) phosphine and incubated at room temperature for 0.5 h. Iodoacetamide (IAA, 6.25 mM) was added to alkylate the protein for 0.5 h at room temperature at dark. The mixture was diluted with 6 volumes of 50 mM ammonium bicarbonate buffer and digested using sequence modified trypsin (1:100 w/w, Promega) for 12 hours at 37 °C.
Peptide desalting
The digested protein solution was quenched with 1 µL phosphoric acid and the pH was adjusted to 2. Then the acidified mixture was slowly loaded onto a 96-well cartridge (Waters, UK) and washed 3 times with 200 µL 0.1% formic acid. After that, the desalted peptides were eluted with 600 µL 50% acetonitrile (in water) and then dried under vacuum (in a speedvac).
LC-MS/MS analysis
The dried peptides were dissolved in 0.1% formic acid (0.5 µg/µL). iRT kit (Biognosys, Switzerland) was added according to manufacturer’s instruction prior to LC-MS/MS analysis. LC-ESI-MS/MS was performed by coupling a nanoLC (Dionex Ultimate 3000, ThermoFisher Scientific, Waltham, USA) to an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific). For each analysis, 2 µL of dissolved peptides was delivered to an analytical column (Dikma, inspire C18, 3 µm, Canada, 150 mm × 75 µm, self-packed) and separated using an 80-minute gradient from 7% to 35% of solvent B (0.1% formic acid in acetonitrile) at 300 nL/minute flow rate.
The Orbitrap Fusion mass spectrometer was operated in data dependent mode, automatically switching between MS and MS/MS. Full scan MS spectra (350-1,550 m/z) were acquired in the Orbitrap at 120,000 resolutions (at m/z 400) after accumulation precursor ions to a target value of 200,000 for a maximum time of 100 ms. Internal lock mass calibration was performed using the ion signal (Si(CH3)2O)6 H+ at m/z 445.120025 present in ambient laboratory air. Tandem mass spectra were recorded for maximum 3 seconds by higher energy collision induced dissociation (HCD, target value of 10,000, max 35 ms accumulation time) at a normalized collision energy of 30% in the ion trap. To maximize the number of precursors targeted for analysis, dynamic exclusion was enabled with one repeat count in 60 s exclusion time.
Peptide and protein identification and quantification
Peaks lists were generated from raw MS data files and were then searched against Uniprot Human Protein Database using Proteome Discoverer (version 1.4, ThermoFisher Scientific). The search was performed considering carbamidomethylation of cysteine residues as fixed modification and methionine oxidation as variable modifications. Trypsin was specified as the proteolytic enzyme and up to two missed cleavages were allowed. The mass tolerances were set to 10 ppm for the precursor ions and 0.06 Da for the fragments. All peptides were filtered at high confidence level provided by the software. Further data interpretation was performed using DAVID, v6.8 and R.
Results
To explore for optimized lysis buffers and conditions, we carefully designed three groups of experimental parameters (see Figure 1 for workflow and Table 2 for buffer composition and condition) for the three types of tissues, which resulted in eleven lysis experimental settings in total [in normal thymus tissue, 3 acid-labile surfactant (ALS) concentrations were tested, see detail in Table 1].

Full table
Comparison of different lysis buffers
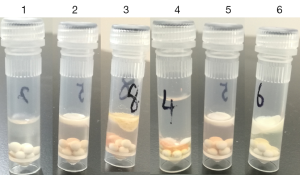
First of all, five pieces of thymus tissue (Normal) with equal masses (80 mg) were chosen to be lysed under the corresponding conditions (Table 1). Considering the high adipose composition of thymus tissue, N1 was lysed in buffer A followed by chloroform/methanol extraction and acetone precipitation. To sufficiently extract proteins from membranes, N2 was designed to be lysed with a slightly modified RIPA buffer followed by acetone precipitation to remove detergents which are not compatible with the downstream proteomics analysis. N3-N5 were used to examine for the optimal concentration of ALS in the lysis buffer. Since ALS can degrade easily under acidic conditions, it is preferred to be used in proteomics analysis. Direct observations after homogenization showed that under protocol B (see Figure 2, tube 1–3), the upper lipid layers of para-tumor and normal thymus tissue after centrifugation were much thicker than thymomas, which is in accordance with their adipose dominant feature. And for para-tumor tissue (tube 4–6), chloroform/methanol extraction could significantly remove the lipids than the other two protocols. Protein amounts obtained from each tissue type using corresponding protocol were summarized in Table 1. As expected, buffer A in together with chloroform/methanol extraction showed inadequate in extracting thymus proteins, with a very low protein yield (40 µg in total). This is probably because the buffer applied is not strong enough to lyse the tissues. On the other hand, strong detergents applied in Protocol B resulted in a significant improvement of protein extraction, with an overall protein amount of 1,268 µg which is nearly 30 times of Protocol A. However, after acetone precipitation, protein amount was significantly reduced to 217.5 µg. Consequently, the RIPA buffer approach appears not desirable. Overall, the employment of acetone precipitation in both protocol A and B had led to a significant protein loss (3–5 times), therefore we went on exploring for a detergent-free, simple but efficient protocol. Acid labile surfactants were reported to have good membrane breaking capabilities and degrade easily under acidic conditions (25), therefore we tested the ability of ALS to facilitate membrane breaking and protein extraction. N3–N5 were designed with a simplified, one-step procedure, in which only different concentrations of ALS (2%, 0.4%, 0.2%) were compared. As shown in Table 1, the protein extraction efficiency was positively correlated with the concentration of ALS. Therefore, application of 2% of ALS resulted in the highest protein amount of 996 µg, in comparison with the total protein amount of 700 and 376 µg obtained in the 0.4% and 0.2% of ALS applications, respectively. Then we chose 2% ALS for protocol C in the experiments in evaluating the para-tumor and thymoma tissues. In a quite similar manner, protocol A completely failed in extracting proteins from the para-tumor tissues, suggesting a requirement for detergents/surfactants in such tissue lysis. Protocol B again obtained the highest amount of proteins (nearly 2 times than protocol C). However, the following acetone precipitation applied enormously reduced total protein amount, ending up with only about 20% amount of total protein harvested by protocol C. Finally, in evaluating the three protocols on thymoma tissue, protocol A again resulted in very low protein quantity. Surprisingly, protocol C gave 2 times more proteins than protocol B before the precipitation step, and eventually provided 6 times more protein quantity than protocol B after precipitation. Consequently, protocol C outperformed the other two protocols in all three analyzed tissues.
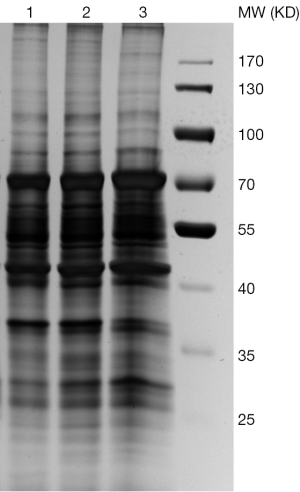
We then went on to visualize the differences of extracted proteins by silver-staining SDS-PAGE (See Figure 3). Again, all the three types of tissues are compared, namely T (Thymoma), P (Para-tumor) and N (Normal). The first lane in each sample, e.g., T1, P1, N1, represented proteins obtained via protocol A, in which only very few protein bands were detected. The lanes 2 and 3 showed the proteins extracted using modified RIPA buffer before and after acetone precipitation (equal loading amount), respectively. These two bands look quite similar from each other. The lane 4 displayed the proteins harvested from protocol C using 2% ALS.
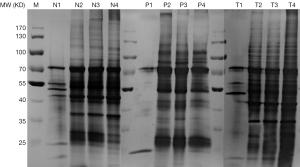
Optimization of ALS concentrations
We then further examined the optimal amount of this surfactant that is required to sufficiently lyse the tissues. Three concentrations of ALS were tested (see Figure 4) and the results were visualized by silver-stained SDS-PAGE. Although using the higher concentration of ALS (2%) did result in a higher total protein yield (Table 1), the variety of proteins as shown the SDS-PAGE remained the same even when ALS is 10 times diluted in the lysis buffers. Therefore, we chose 0.2% ALS in our protocol C as a standard procedure in our proteomics analysis.
Proteomic profiling of thymus and thymoma tissues
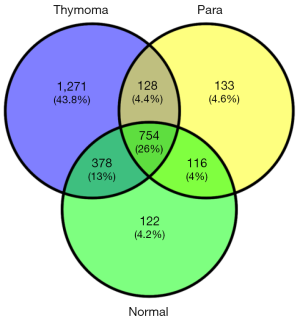
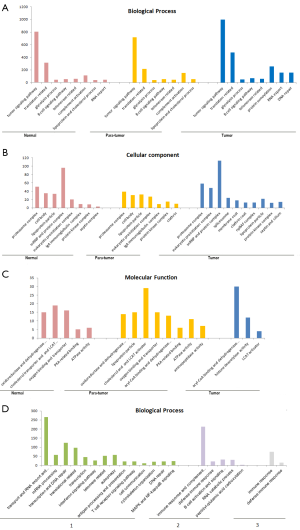
Proteomic profiling analysis using LC-MS/MS of the three tissue types showed that there are a total of 2,902 proteins identified from all the three tissue types (see Figure 5), in which 754 proteins were identified in all three samples. Thymoma tissue provided the highest diversity of proteins, with a total identification of 2,531 proteins, in which 1,271 proteins (43.8% of overall protein IDs) are uniquely identified. This is probably due to the heterogeneous nature of thymoma tissue compared to thymus and para-tumor tissue, in which only 1,370 and 1,131 proteins were identified, respectively. Thymus and para-tumor tissue shared many histological similarities. Therefore 870 proteins were identified in both cases. There are additionally 261 proteins identified in para-tumor tissue, indicating the tumour microenvironment could influence the protein profiles of cancer adjacent tissues.
GO analysis of the identified proteins also provided a lot of information which was not available before. The majority of identified proteins between para-tumor and normal tissue were shared, since the cellular components of these tissues were quite close to each other. Proteins involving in complement activations found in both thymus and para-tumor tissue are a lot more than in thymoma tissues (see Figure 6). Meanwhile, in the analysis of proteins that were identified only from thymus/para-tumor, immune response is the most enriched biological process. This reflects that despite thymic involution in adults, the function of thymus as an immune organ was still partially activated. In addition, due to the adipose dominant feature of the normal/para-tumor tissue, the presence of abundant lipoprotein microparticles contributed to the enrichment lipid/cholesterol metabolism related enzymes, these proteins were however not obviously enriched in thymoma tissues.
Discussion
We applied three representative tissue types (workflow see Figure 1), namely Normal (thymus), Thymoma and Para-tumor. These three tissue types can provide informative and complementary proteomic profiles which facilitate in-depth understanding of the mechanism involved in carcinogenesis from thymus tissue.
Proteomic analysis is a powerful method for analyzing heterogeneous tissue samples such as thymus and thymic malignancies. Owing to its adipose dominant feature of thymus, efficient protein isolation remains a major challenge for MS-based proteomics analysis. It is therefore helpful to establish a suitable sample preparation protocol with effective protein extraction and digestion to peptides followed by LC-MS/MS. Here, we took eleven thymus/thymoma tissue samples (three different types) and optimized five different lysis conditions in terms of protein extract efficiency and specificity, results were further confirmed by SDS-PAGE and LC-MS/MS.
The chloroform/methanol extraction could nicely enrich the lipids and their related proteins, thus is a good choice for this purpose during sample preparation. The variety of proteins obtained using RIPA and ALS had a lot of similarities. However, the introduction of protein precipitation step in protocol B led to a significant loss of protein quantity. Besides, other detergent removal digestion methods, e.g., in-gel digestion, would also introduce more risks such as contaminations and more handling steps would bring more challenges for the quantitative reproducibility in large scale clinical sample preparation practices.
By analyzing proteins that were only identified from thymoma tissues, we found that many proteins obtained from this thymoma sample are related to the processes of DNA repair and protein sumoylation. Functional analysis suggested an increased activity of histone deacetylase in thymoma, while the cellular compartment of the same group of proteins were mainly located in chromatin and spliceosome. This implied that the carcinogenesis of thymus might be closely related to chromatin modifications and DNA repair. Meanwhile, T-cell receptor and IFN mediated pathways were also found to be obviously enriched in thymomas, which also suggested that thymic carcinogenesis could also be associated with the altered immune status of patients. However, it must be noted that thymomas are a series of highly heterogeneous diseases with complex histological and genetic background. Therefore, more samples from other histological types (e.g., Type A, AB, B1, B3) must be further involved in order to comprehensively understand the mechanism of this disease.
Conclusions
Protein levels in tissue samples that differ at different physiological states and/or anatomic locations can be reliably and accurately determined by MS based proteomics analysis. In this work we specifically addressed the sample preparation issues in the thymus and thymoma tissues that limited them from successful proteomics analysis due to the adipose tissue dominant feature. By comparison of 11 experimental conditions, a suitable proteomics tissue sample preparation protocol was optimized and presented here that can facilitate future proteomics analysis on adipose-rich samples. More importantly, using this protocol, we identified more than 2,500 proteins from human thymoma tissues and 1,000 proteins from human thymus tissues. Further analysis on the molecular functions and cellular locations of such proteomes would provide a significantly improved base towards better understanding on mechanisms of thymus carcinogenesis, possible recurrence, and metastasis.
Acknowledgments
The authors want to sincerely thank Dr. Lei Zhu and Dr. Keke Yu, who provided pathological expertise to verify tissue histotypes. We also want to thank Jie Xing, for the technical assistance in the biobank.
Funding: This work is supported by grants from the National Key Research and Development Program of China (No. 2016YFC0902100), the National Natural Science Foundation of China (No. 21708024), Shanghai Sailing Program (No. 16YF1406400), Shanghai Municipal Natural Science Foundation (No.17ZR1426600), Medical Engineering Cross Fund of Shanghai Jiao Tong University (No. YG2016MS80) and Shanghai Jiao Tong University, College of Medicine Biobank Fund (YBKL2013009).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.35). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethical and Protocol Review Committee of Shanghai Chest Hospital, with the ethical approval ID number of KS1615. All procedures performed in studies were in accordance with the ethical standards of Shanghai Chest hospital. Written informed consents were obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zdrojewicz Z, Pachura E, Pachura P. The Thymus: A Forgotten, But Very Important Organ. Adv Clin Exp Med 2016;25:369-75. [Crossref] [PubMed]
- Burger T, Schaefer IM, Kuffer S, et al. Metastatic type A thymoma: morphological and genetic correlation. Histopathology 2017;70:704-10. [Crossref] [PubMed]
- Wang Z, Li H, Cao H, et al. Clinicopathological features of type AB thymoma with liver metastases. Int J Clin Exp Pathol 2014;7:8700-5. [PubMed]
- Kim SH, Koh IS, Minn YK. Pathologic Finding of Thymic Carcinoma Accompanied by Myasthenia Gravis. J Clin Neurol 2015;11:372-5. [Crossref] [PubMed]
- Wang W, Liu D, Yang L, et al. CXCR4 overexpression correlates with poor prognosis in myasthenia gravis-associated thymoma. Hum Pathol 2016;49:49-53. [Crossref] [PubMed]
- Conti S, Gallo E, Sioletic S, et al. Molecular genetic alterations in egfr CA-SSR-1 microsatellite and egfr copy number changes are associated with aggressiveness in thymoma. J Thorac Dis 2016;8:386-95. [Crossref] [PubMed]
- Huang PW, Chang KM. Solitary metastasis to the breast after complete resection of encapsulated type AB thymoma: a case report. J Med Case Rep 2015;9:63. [Crossref] [PubMed]
- Li SY, Wang YX, Wang L, et al. Cytoplasm estrogen receptor beta5 as an improved prognostic factor in thymoma and thymic carcinoma progression. Oncol Lett 2015;10:2341-6. [Crossref] [PubMed]
- Watanabe N, Umemura S, Niho S, et al. Docetaxel for platinum-refractory advanced thymic carcinoma. Jpn J Clin Oncol 2015;45:665-9. [Crossref] [PubMed]
- Yokoyama S, Miyoshi H, Nishi T, et al. Clinicopathologic and Prognostic Implications of Programmed Death Ligand 1 Expression in Thymoma. Ann Thorac Surg 2016;101:1361-9. [Crossref] [PubMed]
- Bellissimo T, Russo E, Ganci F, et al. Circulating miR-21-5p and miR-148a-3p as emerging non-invasive biomarkers in thymic epithelial tumors. Cancer Biol Ther 2016;17:79-82. [Crossref] [PubMed]
- Berardi R, Brunelli A, Pagliaretta S, et al. Impact of VEGF, VEGFR, PDGFR, HIF and ERCC1 gene polymorphisms on thymic malignancies outcome after thymectomy. Oncotarget 2015;6:19305-15. [Crossref] [PubMed]
- Du MJ, Shen Q, Yin H, et al. Diagnostic roles of MUC1 and GLUT1 in differentiating thymic carcinoma from type B3 thymoma. Pathol Res Pract 2016;212:1048-51. [Crossref] [PubMed]
- Keijzers M, Rensspiess D, Pujari S, et al. Expression of pRb and p16INK4 in human thymic epithelial tumors in relation to the presence of human polyomavirus 7. Diagn Pathol 2015;10:201. [Crossref] [PubMed]
- Pilch H, Hohn H, Freitag K, et al. Improved assessment of T-cell receptor (TCR) VB repertoire in clinical specimens: combination of TCR-CDR3 spectratyping with flow cytometry-based TCR VB frequency analysis. Clin Diagn Lab Immunol 2002;9:257-66. [PubMed]
- Radovich M, Solzak JP, Hancock BA, et al. A large microRNA cluster on chromosome 19 is a transcriptional hallmark of WHO type A and AB thymomas. Br J Cancer 2016;114:477-84. [Crossref] [PubMed]
- Zhang T, Chen XU, Chu X, et al. Slug overexpression is associated with poor prognosis in thymoma patients. Oncol Lett 2016;11:306-10. [Crossref] [PubMed]
- Addona TA, Shi X, Keshishian H, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol 2011;29:635-43. [Crossref] [PubMed]
- Latterich M, Schnitzer JE. Streamlining biomarker discovery. Nat Biotechnol 2011;29:600-2. [Crossref] [PubMed]
- McIntosh M, Fitzgibbon M. Biomarker validation by targeted mass spectrometry. Nat Biotechnol 2009;27:622-3. [Crossref] [PubMed]
- Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer 2016;16:525-37. [Crossref] [PubMed]
- Whiteaker JR, Lin C, Kennedy J, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol 2011;29:625-34. [Crossref] [PubMed]
- Borrebaeck CA. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat Rev Cancer 2017;17:199-204. [Crossref] [PubMed]
- Sajic T, Hopfgartner G, Szanto I, et al. Comparison of three detergent-free protein extraction protocols for white adipose tissue. Anal Biochem 2011;415:215-7. [Crossref] [PubMed]
- Ross AR, Lee PJ, Smith DL, et al. Identification of proteins from two-dimensional polyacrylamide gels using a novel acid-labile surfactant. Proteomics 2002;2:928-36. [Crossref] [PubMed]