
Methylation of O6-methylguanine DNA methyltransferase promoter is a predictive biomarker in Chinese melanoma patients treated with alkylating agents
Introduction
Melanoma is a highly aggressive skin cancer with high treatment resistance. Although the U.S. Food and Drug Administration (FDA) has approved various targeted therapies for melanoma which includes BRAF and MEK mutation inhibitors, based on the improved responses observed among patients harboring such mutations, many patients acquire therapeutic resistance and therefore do not achieve persistent, long-term responses (1). Antibodies specific for immune checkpoints, such as those targeting CTLA-4 or the PD1/PDL1 interaction, have also been approved by the FDA and have become a standard of care for patients with unresectable or metastatic melanoma; however, these therapeutic antibodies provide benefits to only some patients (2).
Despite the above limitations, chemotherapy remains a useful first- or second-line treatment for advanced metastatic melanoma (3). Alkylating chemotherapeutic agents such as temozolomide (TMZ) and dacarbazine (DTIC), which are commonly used to treat melanomas, cause DNA double-strand breaks, subsequent termination of replication, and apoptosis in tumor cells (4,5). However, a previous study of TMZ monotherapy reported an extremely objective response rate (13–15%) and median overall survival (OS) (7.7–8.4 months) (6). These findings underscore the need to explore the chemorefractory mechanisms exploited by melanomas and to identify biomarkers that can predict the prognosis of patients treated with alkylating agents. Moreover, a better understanding of biomarkers might provide new combination therapy strategies.
The O6-methylguanine-DNA methyltransferase (MGMT) pathway is considered highly significant among the mechanisms of resistance to alkylating agents. MGMT is a DNA repair enzyme encoded by the MGMT gene at locus 10q26 (7). Largely anecdotal evidence suggests that MGMT plays a central role in preventing the transformation of a physiologic proliferation into malignancy (8). In addition to protecting normal cells from carcinogenesis, MGMT activity also protects tumor cells from the lethal effects of alkylating agents by removing methyl groups from the O6 position of guanine (9).
During the oncogenesis of several human cancers, MGMT was found to be transcriptionally silenced by promoter hypermethylation. The immediate consequence of this phenomenon is the loss of MGMT protein expression and a reduced DNA repair capacity, which results in increased sensitivity to alkylating agents and decreased tumor survival (10,11). Therefore, several studies have described the MGMT promoter methylation status as a predictive biomarker of drug efficacy and prognosis, especially among patients with glioblastoma (GBM) (12). For example, a phase III clinical trial conducted by the European Organization for Research and Treatment of Cancer (EORTC) and National Cancer Institute of Canada (NCIC) identified MGMT methylation as the strongest predictor of TMZ chemotherapy outcomes in patients with GBM; specifically, MGMT methylation was beneficial in terms of response, progression-free survival, and OS (12). Several additional studies have found associations of MGMT inactivation with improved outcomes in Caucasian patients with melanoma who were treated with alkylating agents (13-15). However, as Caucasian and non-Caucasian patients exhibit significant differences in terms of pathogenesis and pathologic subtypes (16,17), the predictive role of MGMT should also be determined in a large cohort of non-Caucasian patients with melanoma.
Many previous reports of ovarian, colorectal, and brain cancers, including malignant astrocytomas, have associated MGMT methylation with TP53 mutation, particularly the G:C to A:T transition (5,11,18-20). By contrast, however, other studies have not identified a relationship between these factors (21,22), indicating the need for further investigation of the potential correlation between MGMT promoter methylation and TP53 mutation and the corresponding effect on prognosis in patients with melanoma. Accordingly, this study aimed to investigate the role of MGMT promoter methylation and TP53 mutation as prognostic biomarkers prognosis in Chinese patients with melanoma who were treated with alkylating agents.
Methods
Patients and tumor tissue samples
Archived formalin-fixed, paraffin-embedded (FFPE) melanoma samples were collected from 205 patients (including 64 with mucosal melanomas, 107 with acral melanomas, and 34 with non-acral skin melanomas) who received DTIC/TMZ and cisplatin-based chemotherapy at the Peking Cancer Hospital & Institute (Beijing, China) between 2005 and 2016 were collected for this study. The samples were analyzed by hematoxylin and eosin staining and immunohistochemistry to confirm the diagnosis of melanoma. Clinical case data were also collected, including patient age and sex, TNM (tumor-node-metastasis) stage, thickness (Breslow), ulceration, and survival outcomes (for which patients were followed until loss to follow-up or death). This study was approved by the Medical Ethics Committee of the Beijing Cancer Hospital & Institute (2017KT21) and was conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants.
DNA isolation and bisulfite modification
Genomic DNA was isolated using the Omega FFPE DNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA). Subsequently, the isolated genomic DNA was subjected to sodium bisulfite modification using the MethylampTM DNA Modification Kit (EpiGentek Inc., Farmingdale, NY, USA) according to the manufacturer’s protocol. The bisulfite-treated DNA was stored at −80 °C.
Detection of MGMT methylation
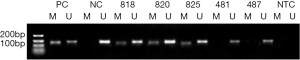
In each tumor sample, the methylation status of the MGMT promoter was determined using methylation-specific polymerase chain reaction (MS-PCR). Bisulfite-treated DNA was amplified using primers specific to either modified or unmodified DNA. The following primer sequences based on previous research (23,24) were used: 5'-TTTCGACGTTCGTAGGTTTTCGC-3' (forward) and 5'-GCACTCTTCCGAAAACGAAACG-3' (reverse) for the methylated reaction and 5'-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3' (forward) and 5'-AACTCCACACTCTTCCAAAAACAAAACA-3' (reverse) for the unmethylated modified reaction. The unmethylated reaction yielded a 91-base pair (bp) product, and the methylated reaction yielded an 81-bp product that included the PYRCpG3, PYRCpG4, and PYRCpG5 region. The PCRs were performed in total volume of 25 µL, and tumor DNA was amplified during 40 cycles at an annealing temperature of 66 °C. The A549 cell line, which is known to exhibit MGMT methylation, was used as a control for methylated MGMT, and DNA from the peripheral blood leukocytes of a healthy individual was used as a control for unmethylated MGMT. Template-free H2O was used as a negative PCR control. The PCR products were analyzed by 3% agarose gel electrophoresis and ethidium bromide staining.
MGMT protein immunohistochemistry
FFPE tissue sections were examined by IHC with the monoclonal mouse anti-human MGMT antibody clone MT3.1 (Abcam, Cambridge, UK). The primary antibody was omitted from negative controls. Inflammatory cells and reactive stromal cells served as internal positive controls. Prior to staining, the tissue sections were deparaffinized with xylene for 30 min and rehydrated in decreasing concentrations of ethanol. Endogenous peroxidases were then blocked with 30% H2O2 for 20 min in phosphate-buffered saline (PBS). For antigen retrieval, the slides were heated in 0.01 M citrate buffer (pH 6.0) in a pressure cooker for 4 min and subsequently cooled to room temperature in the same buffer. After washing, the slides were incubated overnight with the primary antibody at 4 °C (dilution 1:100). The tumor sections were then stained using the EnvisionTM Detection kit (Gene Tech, Shanghai, PR China) according to the manufacturer’s instructions and counterstained with hematoxylin. MGMT status was determined based on staining intensity {negative (0), low positive [1], strongly positive [2]} and percentage of positive tumor cells {0% (0), 1–50% [1], 51–100% [2]}. Only nuclear staining was regarded as positive staining.
TP53 mutation detection
Exons 5–8 of TP53 were amplified by a two-step PCR. The following primer sequences were used for step I amplification: 5'-GTTTCTTTGCTGCCGTCTTC-3' (forward) and 5'-CCTTCCACTCGGATAAGATG-3' (reverse) for exon 5, 5’-AGCACATGACGGAGGTTGTG-3' (forward) and 5'-TCTCATGGGGTTATAGGGAG-3' (reverse) for exon 6, 5'-GCCTCCCCTGCTTGCCACAG-3' (forward) and 5'-GAGAGGTGGATGGGTAGTAG-3' (reverse) for exon 7, and 5'-TACCTGGAGCTGGAGCTTAG-3' (forward) and 5'-GAAAGAGGCAAGGAAAGGTG-3' (reverse) for exon 8. The following primer sequences were used for step II amplification: 5'-CTTTATCTGTTCACTTGTGC-3' (forward) and 5'-CAATCAGTGAGGAATCAGAG-3' (reverse) for exon 5, 5'-CATGAGCGCTGCTCAGATAG-3' (forward) and 5'-TAGGGAGGTCAAATAAGCAG-3' (reverse) for exon 6, 5'-CATCTTGGGCCTGTGTTATC-3' (forward) and 5'-GAAGAAATCGGTAAGAGGTG-3' (reverse) for exon 7, and 5'-GACAGGTAGGACCTGATTTC-3' (forward) and 5'-AAGTGAATCTGAGGCATAAC-3' (reverse) for exon 8.
Clinical data analysis and statistical methods
OS was defined as the time from diagnosis to the death of the patient. For the patients who were still alive at the time of the study, OS was defined as the time from diagnosis to the current date. The patients’ clinicopathological characteristics were correlated with the MGMT promoter methylation status and MGMT protein expression, and the significance of these associations was determined using Pearson’s chi-square test, the continuity correction test, Fisher’s exact test, and the linear-by-linear association test as appropriate. Survival data were used to generate Kaplan-Meier curves, which were compared according to MGMT promoter methylation status using the log-rank test. A Cox regression model was used for both univariate and multivariate analyses. Age, sex, primary site, stage, MGMT promoter methylation, MGMT protein expression, TP53 mutation status, and the combination of MGMT promoter methylation and TP53 mutation status were included in the multivariate model, which used a stepwise method for variable selection. The level of statistical significance was set at P<0.05, and all tests were two-sided. The statistical analysis was performed using IBM SPSS statistical software (version 20.0; IBM, Inc., Armonk, NY, USA).
Results
Correlation of MGMT promoter methylation with clinicopathological features
Using MS-PCR, we found that the MGMT promoter was methylated in 97 (47%) of 205 tested melanomas (Table 1 and Figure 1). We next used the Pearson method to evaluate the associations of MGMT promoter methylation with different clinicopathological features (Table 1), and observed a significant association with tumor stage (P=0.017), but not with sex (P=0.735), age (P=0.647), or the primary tumor site (P=0.875).
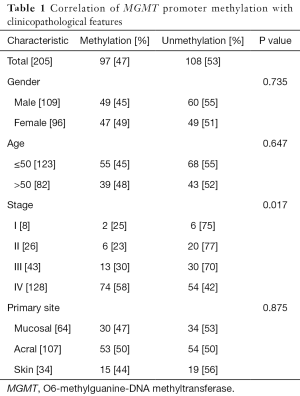
Full table
Association of MGMT protein expression with MGMT promoter methylation
We next used IHC to detect positive nuclear staining for the MGMT protein in 15 of 97 cases with MGMT promoter methylation and 42 of 108 cases without methylation (Table 2). As described in the Materials and Methods, IHC staining was scored from 0 to 2 on the basis of the staining intensity and percentage of positive tumor cells (Figure 2). Strongly positive nuclear MGMT protein staining (IHC score =2) was detected in 8 of 97 patients with methylation and 25 of 108 patients without methylation, whereas low positive nuclear staining (IHC score =1) was detected in 25 of 97 cases with methylation and 17 of 108 cases without methylation.
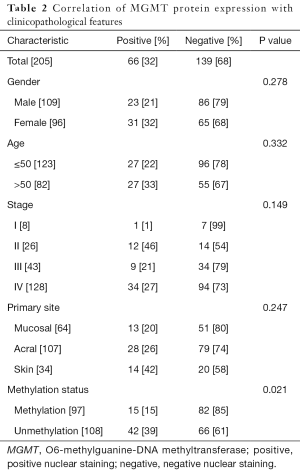
Full table

Using Pearson’s chi-square test, we identified a significant association between MGMT expression and MGMT promoter methylation in patients with melanoma (P=0.021). Notably, both strongly positive and weakly positive nuclear MGMT protein expression were significantly associated with the MGMT promoter methylation status in our melanoma samples (P=0.001 and 0.027, respectively). However, we observed no significant associations of MGMT protein expression with sex (P=0.278), age (P=0.332), stage (P=0.149), or primary tumor site (P=0.247).
Correlation of MGMT promoter methylation with TP53 mutation
To evaluate the potential association of MGMT promoter methylation with TP53 mutation in melanoma, we subjected DNA samples from the same 205 tumors to PCR followed by Sanger sequencing to examine TP53 mutations in exons 5–8, which are commonly affected. We identified 37 cases with TP53 mutations. Of these, the MGMT promoter was methylated in 29 cases (30% of 97 samples with methylation) and unmethylated in only 8 (7% of 108 samples without methylation) (Table 3). Accordingly, we found a significant association of MGMT promoter methylation with TP53 mutation (P=0.004), with an odds ratio of 4.463 [95% confidence interval (CI): 1.879–22.664] When separating the patient by the primary site (mucosal vs. acral vs. skin), there is a significant correlation between MGMT promoter methylation status and TP53 mutation (P=0.01).
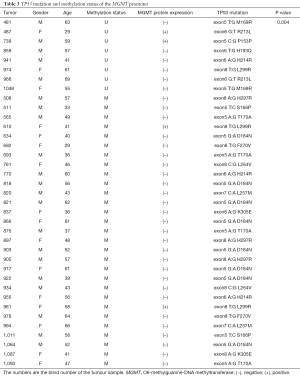
Full table
A further analysis revealed that the melanomas harbored different TP53 mutation types, such as T:G, A:G, and C:G. The G:C to A:T mutation at exon 5 was identified in 8 of 97 (8%) cases with a methylated MGMT promoter and none of the cases with an unmethylated MGMT promoter, indicating that this type of mutation tends to occur under conditions of MGMT promoter methylation (P<0.01). Moreover, the G:C to A:T mutation at exon 5 was detected in 8 of 148 (5%) patients with MGMT protein expression and none of the cases without MGMT protein expression, indicating an increased tendency of this mutation to occur in melanomas expressing MGMT protein. However, this latter association was not statistically significant (P=0.109).
Association of MGMT promoter methylation and TP53 mutation with OS among patients with melanoma following treatment with alkylating agents
In our univariate Cox analyses, we identified stage and MGMT promoter methylation as factors significantly associated with OS among melanoma patients treated with alkylating agents (Table 4). The multivariate Cox regression model subsequently identified both factors as significantly predictive of OS (Table 4).

Full table
The median OS time of all 205 patients was 38.9 months, and a Kaplan-Meier analysis showed that patients with MGMT promoter methylation had a significantly greater median OS, compared to those without MGMT promoter methylation (36.7 vs. 23.1 months, P=0.021, Figure 3A). Although patients without MGMT protein expression also had a greater OS than did those with MGMT protein expression, this difference was not significant (34.5 vs. 25.9 months, P=0.092). Similarly, when patients were grouped by MGMT IHC staining scores, those with scores of 0 had poorer OS relative to those with scores of 1 or 2, although this difference was not statistically significant (36.7 vs. 26.1 and 21.5 months, respectively, P=0.086, Figure 3B).
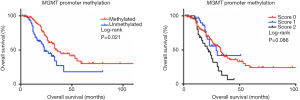
Although patients harboring a TP53 mutation or the combination of a TP53 mutation and MGMT promoter methylation had a relatively better OS outcome, this difference was also not significant (33.4 vs. 28.5 months, P=0.247 and 35.1 vs. 27.2 months, P=0.259, respectively). Finally, the stratification of patients by primary tumor site (mucosal vs. acral vs. skin) revealed that patients without MGMT promoter methylation exhibited worse OS, compared to those with MGMT promoter methylation (P=0.015).
Discussion
Although the DNA alkylating agents TMZ and DTIC are considered standard components of chemotherapy for melanoma, patients who receive TMZ for stage IV melanoma have a low response rate (13–15%) and dismal prognosis (OS, 7.7–8.4 months) (6). In addition, only 5% of patients achieve a complete response with TMZ or DTIC treatment, and few achieve a sustained response to either drug (25,26). However, the factors that might better identify patients who may benefit from TMZ or DTIC chemotherapy remain largely unknown. Therefore, in this study, we aimed to evaluate the prognostic role of MGMT promoter methylation and TP53 mutation in a large cohort of Chinese patients treated with alkylating agents for melanoma. In our population of 205 patients, we observed a MGMT promoter methylation rate of 47%, which was demonstrably higher than the corresponding frequency among Caucasian patients (21.5–26%) (15,27). This discrepancy may reflect differences in the pathogenesis and pathological subtypes of melanoma between non-Caucasians and Caucasians.
Additionally, our analysis of correlations between MGMT promoter methylation and clinicopathological features found that MGMT methylation only associated significantly with tumor stage, but not with sex, age, or primary tumor site. Specifically, MGMT promoter methylation was significantly more frequent among high-grade (stages III, IV) melanomas, compared to low-grade (I, II) melanomas (51% vs. 24%, P=0.004). However, we note that the large difference in the numbers of low-grade and high-grade melanomas (34 vs. 171) included in this analysis might have led to statistical bias. Still, our findings are consistent with a previous study, which found that MGMT promoter methylation was more common in advanced-stage tumors relative to early-stage tumors (28). Taken together, these findings suggest that MGMT promoter methylation and consequent inactivation may play a crucial role in melanoma progression.
Immunohistochemistry revealed a lack of MGMT expression in 82 of 97 (85%) tumors exhibiting MGMT promoter methylation, compared to 66 of 108 (61%) tumors with an unmethylated MGMT promoter. This finding indicates differences in MGMT protein expression between melanomas that do and do not exhibit MGMT promoter methylation (P=0.021); specifically, promoter methylation is associated with a loss of protein expression. However, the high percentage of melanomas lacking both MGMT promoter expression and detectable MGMT protein expression suggests that the latter process may be regulated by additional factors. Additionally, technical issues and other factors, such as tumor heterogeneity, may have influenced the results, as a previous study indicated that the use of different methods to assess the MGMT promoter methylation status might affect the results (29). In a phase II study of extended-dose temozolomide in patients with melanoma, Petra performed MS-PCR to detect the MGMT promoter methylation rate of 26%, and Rainer and his colleague determined a MGMT promoter methylation frequency of 21.5% via methylation-sensitive high-resolution melting (15,27). In our cohort, we used MS-PCR to detect the MGMT promoter methylation status. Petra reported that IHC staining for MGMT could not predict the responses of melanoma patients treated with TMZ (30). However, Augustine profiled the expression of 38,000 genes using an oligonucleotide-based microarray platform, and found a significant correlation of MGMT expression with TMZ sensitivity in the context of melanoma (P≤0.0001) (31). In our cohort, we used IHC to evaluate the expression of MGMT protein in melanomas but found that this factor could not predict the prognosis of patients treated with alkylating agents. Therefore, future studies should focus on gene expression profiling. Regarding tumor heterogeneity, promoter hypermethylation heterogeneity has been frequently observed in melanomas and glioblastomas (32,33). However, we could not explore the issue of heterogeneity in this study because of an inability to acquiring tumor tissues from in multiple sites in the body.
We further investigated the correlation of TP53 mutation with MGMT expression, as such correlations have been previously identified in different cancer types, including melanoma and glioblastoma (34-36). In our cohort, we identified TP53 mutations in 18% of melanoma cases, and observed that this mutation was more frequent among patients with MGMT promoter methylation (30%) than among those without MGMT promoter methylation (7%). Several previous studies have indicated an association between MGMT promoter hypermethylation and G:C to A:T transition mutations of the TP53 gene in nervous system tumors and glioblastoma (37,38). Consistent with these earlier findings, we observed a significant incidence of TP53 G:C to A:T mutation in patients with MGMT promoter methylation, (8 of 97, 8%, P<0.01). However, the relationship between the MGMT promoter methylation status and TP53 mutation remains under debate (39-43). Our analysis indicated a significant correlation of MGMT promoter methylation with TP53 mutation (P=0.004), suggesting that the former may increase the occurrence of the latter in melanoma. It is therefore reasonable to surmise that MGMT promoter methylation and TP53 mutation are not independent in the carcinogenesis of melanoma. However, the underlying mechanism and functional aspects require further investigation.
We believe that the most crucial finding of the present study is the association of MGMT promoter methylation with a significantly longer OS among patients with melanoma. In our multivariate Cox analyses, we identified the MGMT promoter methylation status as an independent prognostic variable associated with a longer OS. This is the first study to demonstrate the prognostic value of MGMT promoter methylation in non-Caucasian melanoma population treated with alkylating agents.
Although previous studies have established that the MGMT promoter methylation status can predict the efficacy of TMZ treatment in patients with glioblastoma (44,45), randomized clinical trials of a combination regimen comprising the MGMT inhibitor lomeguatrib (LM) and TMZ for metastatic cutaneous melanoma have not demonstrated any significant improvements in patient responses or OS when compared to TMZ monotherapy in a Caucasian population (46). As we noted previously, however, epidemiologic factors differ greatly between Asian and Caucasian populations. The predictive value of MGMT promoter methylation regarding response to alkylating agents needs to be supported by further validation. Moreover, the existing evidence merits additional basic research and clinical trials of MGMT promoter methylation.
In summary, we have demonstrated the prognostic significance of MGMT promoter methylation in melanoma patients treated with alkylating agents. Notably, the presence of MGMT promoter methylation was associated with the occurrence of TP53 mutation in melanoma. Determination of the MGMT promoter methylation status might help to identify melanoma patients who will likely benefit from chemotherapy with alkylating agents.
Acknowledgments
Funding: This work was supported by grants from National Natural Science Foundation of China (81402264), Beijing Municipal Natural Science Foundation (7152033) and Beijing Municipal Administration of Hospitals Clinical medicine Development of special funding support (ZYLX201603).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Medical Ethics Committee of the Beijing Cancer Hospital & Institute (2017KT21) and was conducted according to the principles of the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Iams WT, Sosman JA, Chandra S. Novel Targeted Therapies for Metastatic Melanoma. Cancer J 2017;23:54-8. [Crossref] [PubMed]
- Chuk MK, Chang JT, Theoret MR, et al. FDA Approval Summary: Accelerated Approval of Pembrolizumab for Second-Line Treatment of Metastatic Melanoma. Clin Cancer Res 2017;23:5666-70. [Crossref] [PubMed]
- Legha SS, Ring S, Bedikian A, et al. Treatment of metastatic melanoma with combined chemotherapy containing cisplatin, vinblastine and dacarbazine (CVD) and biotherapy using interleukin-2 and interferon-alpha. Ann Oncol 1996;7:827-35. [Crossref] [PubMed]
- Margison GP, Santibanez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis 2002;17:483-7. [Crossref] [PubMed]
- Kaina B. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenet Genome Res 2004;104:77-86. [Crossref] [PubMed]
- Middleton MR, Grob JJ, Aaronson N, et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol 2000;18:158-66. [Crossref] [PubMed]
- Paz MF, Yaya-Tur R, Rojas-Marcos I, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res 2004;10:4933-8. [Crossref] [PubMed]
- Inno A, Fanetti G, Di Bartolomeo M, et al. Role of MGMT as biomarker in colorectal cancer. World J Clin Cases 2014;2:835-9. [Crossref] [PubMed]
- Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem 1980;255:10569-71. [PubMed]
- Costello JF, Futscher BW, Tano K, et al. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem 1994;269:17228-37. [PubMed]
- Yin D, Xie D, Hofmann WK, et al. DNA repair gene O6-methylguanine-DNA methyltransferase: promoter hypermethylation associated with decreased expression and G:C to A:T mutations of p53 in brain tumors. Mol Carcinog 2003;36:23-31. [Crossref] [PubMed]
- Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 2010;6:39-51. [Crossref] [PubMed]
- Busch C, Geisler J, Lillehaug JR, et al. MGMT expression levels predict disease stabilisation, progression-free and overall survival in patients with advanced melanomas treated with DTIC. Eur J Cancer 2010;46:2127-33. [Crossref] [PubMed]
- Cesinaro AM, Sartori G, Migaldi M, et al. Prognostic significance of MGMT gene promoter methylation in differently treated metastatic melanomas. Pathology 2012;44:313-7. [Crossref] [PubMed]
- Schraml P, von Teichman A, Mihic-Probst D, et al. Predictive value of the MGMT promoter methylation status in metastatic melanoma patients receiving first-line temozolomide plus bevacizumab in the trial SAKK 50/07. Oncol Rep 2012;28:654-8. [Crossref] [PubMed]
- Chi Z, Li S, Sheng X, et al. Clinical presentation, histology, and prognoses of malignant melanoma in ethnic Chinese: a study of 522 consecutive cases. BMC Cancer 2011;11:85. [Crossref] [PubMed]
- Guo J, Si L, Kong Y, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol 2011;29:2904-9. [Crossref] [PubMed]
- Hengstler JG, Tanner B, Moller L, et al. Activity of O(6)-methylguanine-DNA methyltransferase in relation to p53 status and therapeutic response in ovarian cancer. Int J Cancer 1999;84:388-95. [Crossref] [PubMed]
- Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 2001;61:4689-92. [PubMed]
- Watanabe T, Katayama Y, Komine C, et al. O6-methylguanine-DNA methyltransferase methylation and TP53 mutation in malignant astrocytomas and their relationships with clinical course. Int J Cancer 2005;113:581-7. [Crossref] [PubMed]
- Osanai T, Takagi Y, Toriya Y, et al. Inverse correlation between the expression of O6-methylguanine-DNA methyl transferase (MGMT) and p53 in breast cancer. Jpn J Clin Oncol 2005;35:121-5. [Crossref] [PubMed]
- Jesien-Lewandowicz E, Jesionek-Kupnicka D, Zawlik I, et al. High incidence of MGMT promoter methylation in primary glioblastomas without correlation with TP53 gene mutations. Cancer Genet Cytogenet 2009;188:77-82. [Crossref] [PubMed]
- Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000;343:1350-4. [Crossref] [PubMed]
- Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999;59:793-7. [PubMed]
- Serrone L, Zeuli M, Sega FM, et al. Dacarbazine-based chemotherapy for metastatic melanoma: thirty-year experience overview. J Exp Clin Cancer Res 2000;19:21-34. [PubMed]
- Kim C, Lee CW, Kovacic L, et al. Long-term survival in patients with metastatic melanoma treated with DTIC or temozolomide. Oncologist 2010;15:765-71. [Crossref] [PubMed]
- Tuominen R, Jewell R, van den Oord JJ, et al. MGMT promoter methylation is associated with temozolomide response and prolonged progression-free survival in disseminated cutaneous melanoma. Int J Cancer 2015;136:2844-53. [Crossref] [PubMed]
- Lai JC, Cheng YW, Goan YG, et al. Promoter methylation of O(6)-methylguanine-DNA-methyltransferase in lung cancer is regulated by p53. DNA Repair (Amst) 2008;7:1352-63. [Crossref] [PubMed]
- Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 2012;118:4201-11. [Crossref] [PubMed]
- Rietschel P, Wolchok JD, Krown S, et al. Phase II study of extended-dose temozolomide in patients with melanoma. J Clin Oncol 2008;26:2299-304. [Crossref] [PubMed]
- Augustine CK, Yoo JS, Potti A, et al. Genomic and molecular profiling predicts response to temozolomide in melanoma. Clin Cancer Res 2009;15:502-10. [Crossref] [PubMed]
- Rastetter M, Schagdarsurengin U, Lahtz C, et al. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol Histopathol 2007;22:1005-15. [PubMed]
- Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013;369:134-44. [Crossref] [PubMed]
- Naumann SC, Roos WP, Jost E, et al. Temozolomide- and fotemustine-induced apoptosis in human malignant melanoma cells: response related to MGMT, MMR, DSBs, and p53. Br J Cancer 2009;100:322-33. [Crossref] [PubMed]
- Li X, Yuan L, Zhao J, et al. Adenovirus-based strategies enhance antitumor capability through p53-mediated downregulation of MGMT in uveal melanoma. Cancer Biol Ther 2017;18:194-9. [Crossref] [PubMed]
- Lotfi M, Afsharnezhad S, Raziee HR, et al. Immunohistochemical assessment of MGMT expression and p53 mutation in glioblastoma multiforme. Tumori 2011;97:104-8. [Crossref] [PubMed]
- Bello MJ, Alonso ME, Aminoso C, et al. Hypermethylation of the DNA repair gene MGMT: association with TP53 G:C to A:T transitions in a series of 469 nervous system tumors. Mutat Res 2004;554:23-32. [Crossref] [PubMed]
- Wang X, Chen JX, Liu JP, et al. Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann Surg Oncol 2014;21:1337-44. [Crossref] [PubMed]
- Fu XR, Sun ZC, Chang Y. Expression and clinical significance of P53, O6-methylguanine-dna methyltransferase and epidermal growth factor receptor in glioma. J Biol Regul Homeost Agents 2015;29:853-8. [PubMed]
- Pietrantonio F, Perrone F, de Braud F, et al. Activity of temozolomide in patients with advanced chemorefractory colorectal cancer and MGMT promoter methylation. Ann Oncol 2014;25:404-8. [Crossref] [PubMed]
- Ogura R, Tsukamoto Y, Natsumeda M, et al. Immunohistochemical profiles of IDH1, MGMT and P53: practical significance for prognostication of patients with diffuse gliomas. Neuropathology 2015;35:324-35. [Crossref] [PubMed]
- Su Y, Yin L, Liu R, et al. Promoter methylation status of MGMT, hMSH2, and hMLH1 and its relationship to corresponding protein expression and TP53 mutations in human esophageal squamous cell carcinoma. Med Oncol 2014;31:784. [Crossref] [PubMed]
- Liu Y, Wang J, Zhang H, et al. The expression of O6-methylguanine DNA methyltransferase and p53 in non-small cell lung cancer and the association with the prognosis. Zhonghua Jie He He Hu Xi Za Zhi 2010;33:427-31. [PubMed]
- Brandes AA, Tosoni A, Franceschi E, et al. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation With MGMT promoter methylation status. J Clin Oncol 2009;27:1275-9. [Crossref] [PubMed]
- Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol 2013;31:4085-91. [Crossref] [PubMed]
- Ranson M, Hersey P, Thompson D, et al. Randomized trial of the combination of lomeguatrib and temozolomide compared with temozolomide alone in chemotherapy naive patients with metastatic cutaneous melanoma. J Clin Oncol 2007;25:2540-5. [Crossref] [PubMed]


