
Methylated septin 9 gene for noninvasive diagnosis and therapy monitoring of breast cancer
Introduction
Breast cancer is one of the most common cancers and a leading cause of cancer-related deaths in women worldwide (1). Screening tests enabling early detection of breast tumors facilitate cancer treatment at early stages, thereby improving patient prognosis and clinical outcomes. Currently available approaches for breast cancer diagnosis include imaging techniques and biomarkers tests (2). Breast ultrasound and molybdenum target radiography are generally less sensitive, thus often lead to false negative results for early-stage cancer patients (3). Magnetic resonance imaging (MRI) has high sensitivity in detecting early stage cancers; however, the specificity of breast MRI is limited and questionable (4,5). Despite common use of imaging approaches in clinical practice, tumor markers have been of great interest to physicians, which allow to detect early disease and help screening in a cost-effective manner. Several tumor markers have been recommended for breast cancer diagnosis, including carcinoembryonic antigen (CEA), estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Nevertheless, use of these markers for screening and diagnosis has been hindered and limited due to the low sensitivity and lack of specificity (6). Thus, there is an urgent need to develop novel, more sensitive, and ideally noninvasive diagnostic biomarkers to maximize the number of women who participate in screening and to reduce breast cancer-related deaths. Despite the intensive research efforts, currently there is no reliable test that can prognosticate the efficacy of the therapies shortly after the treatments.
Circulating tumor DNA (ctDNA) has been proposed as a screening tool in breast cancer screening tests (7). Because breast cancers develop as the result of accumulation of genetic and epigenetic changes (8), and molecular markers concerning genetic and epigenetic alterations have been evaluated (9). DNA methylation is considered as a major tumor-associated epigenetic change (10), since methylation of tumor suppressor genes can result in gene silencing and thus potentially sustain cancerogenesis (11). For this reason, differences in the methylation status of tumor DNA have been evaluated in breast tumors (12). Septin 9 (SEPT9) is a member of the SEPTIN family involved in cytokinesis and cytoskeletal organization (13), and alterations of SEPT9 gene have been linked to multiple cancers (14). However, only few studies so far have evaluated the methylation of the SEPT9 promoter region as a biomarker for breast cancer detection (15,16).
In the present study, we examined the methylation status of SEPT9_v2 in both plasma samples and tumor tissues collected from Chinese breast tumor patients (malignant and benign tumor). Subsequently, we analyzed the correlation between methylated septin 9 (mSEPT9) status and clinicopathological characteristics as well as molecular classifications of breast cancer patients. We also compared the power of plasma mSEPT9 as a diagnostic biomarker with several often used tumor biomarkers including serum CEA, cytokeratin 19 fragments (CK19 or CYFRA 21-1) and carbohydrate antigen 15-3 (CA15-3). Finally, the application of the plasma mSEPT9 as a follow-up biomarker after breast cancer treatment was evaluated.
Methods
Tissues and plasma specimens
A total of 86 breast tumor tissue samples (59 breast cancer and 27 benign breast tumor patients) confirmed by pathologic examinations were obtained from the needle breast biopsy collections of Liaocheng People’s Hospital between August 2016 and June 2017. No statistic differences were detected in either age or menopausal status (Table S1). These patients had no prior history of cancer-related surgery, chemotherapy, or hormonal therapy. The clinicopathological characteristics and molecular classifications of these patients are summarized in Tables 1,2. Tissue samples were snap-frozen in liquid nitrogen and kept at −196 °C until analyses.
Full table
Full table
A total of 207 plasma samples collected from 80 patients with breast cancer, 27 patients with benign breast tumor, and 100 healthy volunteers. There were no statistic differences among these cohorts in either age or menopausal status (Table S2). The healthy volunteers enrolled as controls had no diagnosed neoplasm in their mammary gland or other organs. The clinicopathological parameters and molecular classifications of these cohorts are summarized in Tables 3,4. Twenty-one follow-up specimens were obtained from breast cancer patients who had been treated with neoadjuvant chemotherapy and surgical treatment.
Full table

Full table
Clinical and pathologic data of the breast cancer patients were collected from the electronic medical records. The TNM cancer staging (T: size or direct extent of the primary tumor; N: degree of spread to regional lymph nodes; M: presence of distant metastasis) was assigned based on the seventh edition of the Cancer Staging Manual of the American Joint Committee on Cancer (17). The efficacy of chemotherapy was evaluated by Miller-Payne grade (MP) classification and response evaluation criteria in solid tumors (RECIST) (18,19).
Analysis of mSEPT9 in tissues and plasma
To test the mSEPT9 in biopsy tissues, DNA was extracted from the tissues using DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s recommendations. Briefly, the samples were first lysed using proteinase K and then loaded onto DNeasymini spin columns. The DNA was centrifuged, washed, and then eluted using elution buffer. All DNeasy-purified DNA samples had A260/A280 ratios of 1.7–1.9, and spectrometry scans showed a symmetric peak at 260 nm.
DNA in plasma was extracted using the EpiproColon Plasma Quick Kit (Epigenomics AG, Berlin, Germany). To evaluate SEPT9 methylation, 10 mL peripheral blood samples were collected with 10 mL K2 EDTA anticoagulant tubes (BD-Plymouth, Oxford, UK). The blood samples were stored at 2 to 8 °C for up to 24 h prior to plasma preparation. Each sample was centrifuged for 12 min at 2,500 rpm at room temperature. Plasma was transferred to a clean 15 mL polypropylene centrifuge tube with conical bottom. The sample was centrifuged again for 12 min at 2,500 rpm. Plasma was transferred into a labeled cryovial and stored at −80 °C. To purify circulating DNA, 3.5 mL plasma was mixed with an equal volume of lysis buffer, and the mixture was then incubated at 15 to 30 °C for 10 min. Then, 90 µL Epi proColon Magnetic Beads (freshly suspended) and 2.5 mL of Absolute Ethanol (for molecular biology, ≥99.5%) were added into the plasma. The mixture was subsequently incubated on a rotator at 15 to 30 °C for 45 min at 15 rpm. The magnetic beads were then washed and DNA eluted into 100 µL elution buffer. This procedure usually yields DNA concentrations at a range between 5.1 to 16.3 ng/mL from plasma samples and 40.2 to 709.7 ng/mL from tissue samples.
The purified DNA was then mixed with 150 µL bisulfite solution and 25 µL Epi proColon Protection Buffer, and then incubated for 45 min at 80 °C. The converted DNA (bisulfite-modified DNA, designated asbisDNA hereafter) was then analyzed by duplex real-time PCR using methylation-specific primers on a 7500 real-time PCR system (Applied Biosystems, California, America) with simultaneous detection of both the methylated SEPT9 DNA and the internal control ACTB (ß-actin) DNA (20). The sequences of primers, blocker, and probes for SEPT9 detection used in methylation-specific PCR amplification were as follows: forward primer, 5'-CCCACCAACCATCATAT-3'; reverse primer, 5'-GTAGTAGTTAGTTTAGTATTTATTTT-3'; blocker, 5'-CATCATATCAAACCCCACAATCAACACACAAC-3'; probe 1, 5'-GTTCGAAATGATTTTATTTAGTTGC-3'; probe 2, 5'-CGTTGATCGCGGGGTTC-3' (GenBank Gene ID: 10801) (21). The thermocycling program was as follows: denaturing at 94 °C for 20 min; 45 cycles of 93 °C for 30 s followed by 55.5 °C for 35 sec and 62 °C for 35 s; and cooling at 40 °C for 5 s.
Analysis of SEPT9_v2 mRNA in tissues
Total RNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue sections (4 µm) using RNeasy FFPE Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Then the cDNA was synthesized using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) kit (Takara, Tokyo, JAP) according to the manufacturer’s instructions. SYBR-Green real-time quantitative PCR analyses for mRNAs of SEPT9_v2 and internal control (β-actin) were performed on a 7500 real-time PCR system (Applied Biosystems, California, America) using the following parameters: 95 °C (30 s), followed by 40 cycles of 95 °C (5 s) and 60 °C (30 s).
Immunohistochemistry (IHC) Staining of SEPT9 protein
Tissue samples were fixed in 10% formaldehyde and embedded in paraffin. Serial 5-µm sections were cut, deparaffinized in xylene, and hydrated through an ethanol series. The sections were then heated at 100 °C for 30 min in citrate buffer (pH 6.0) in a microwave oven to retrieve antigens. After quenching endogenous peroxidase with 3% hydrogen peroxide for 15 min at room temperature, non-specific binding was blocked by incubating the slides with SPlink Detection Kits (ZSGB-BIO, Beijing, China) for 30 min, followed by incubation of the sections with a rabbit polyclonal anti-SEPT9 (1:100) antibody (Abnova, Taipei, Taiwan) at 4 °C overnight.
For IHC, the sections were washed three times with PBS, and then incubated for 1 h with a HRP-conjugated Goat anti-rabbit secondary antibody (Epigentek, Wuhan, China) diluted at a ratio of 1:100. The color on the slides was developed for 2 min using a DAB Chromogen Substrate Kit (Solarbio, Beijing, China), and sections were then counter stained with hematoxylin staining to identify cell nuclei. A scoring system was assigned to evaluate the SEPT9 protein expression levels:, 0 for negative staining, +1 for weak staining, +2 for moderate staining, and +3 for strong staining.
Determination of the status of ER, PR, HER2, Ki67 and p53
The status of ER, PR, HER2, Ki-67 as well as the p53 index were determined by IHC method. The antibodies used were as follows: anti-ER (SP1) rabbit monoclonal antibody (Roche Diagnostic GmbH, Mannheim, Germany), anti-PR (1E2) rabbit monoclonal antibody (Roche Diagnostic GmbH), anti-HER2 (4B5) rabbit monoclonal antibody (Roche Diagnostic GmbH), anti-Ki67 (MIB-1) mouse monoclonal antibody (Maxim, Fuzhou, China), and anti-p53 (DO-7) mouse monoclonal antibody (Maxim). The procedures of IHC were as described above.
Interpretation standard: ER and PR were considered as positive when stained nuclei constituted ≥1% of total nuclei, according to the guideline recommendations (22). The Ki67 scores were expressed as the percentage of positively staining cells among the total number of invasive cells in the scored area (23). Neoplastic lesions in each p53-immunostained slide were confirmed using the corresponding hematoxylin and eosin stained specimens (24). Briefly, HER2 staining in tumor tissues were categorized into four grades (0, 1, 2, and 3), and tumors with scores of 3+ grades were considered to be positive according to the guideline recommendations (25). When HER2-IHC was 2+, HER2-FISH (fluorescence in situ hybridization) assay was performed according to the latest ASCO/CAP guideline.
FISH assays were carried out on paraffinized tissue sections (4-µm) using the PathVysion HER-2 DNA Probe Kit (Abbott Laboratories, Des Plaines, USA). HER2 positivity is defined according to American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) recommendations (25). Sixty cells were routinely scored for HER2 signal (red) and CEP17 signal (green). The mean HER2 and CEP17 signals were used to calculate the final FISH ratio.
Measurements of CEA, CK19 and CA15-3
Serum CEA, CK19 and CA15-3 concentrations were analyzed using an Electrochemiluminescence Immunoassay Kit (Roche Diagnostic GmbH, Mannheim, Germany) according to the manufacturer’s instructions. They were defined as positive when detection results were greater than the following values: 5 U/mL (CEA), 30 U/mL (CA15-3) and 3.3 U/mL (CK19), respectively.
Statistical analyses
All statistical analyses were performed using the SPSS18 software (SPSS, Chicago, America). The quantitative data was expressed as mean ± standard deviation (SD) and analyzed using t-test. Chi-square test or Fisher’s exact test (when appropriate) was applied to evaluate the correlation between clinicopathological features and mSEPT9 status. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic performance of mSEPT9 as a biomarker for differentiating breast cancer from benign breast tumor samples (26). Discriminative power was assessed by the area under a receiver operating characteristic curve (AUC). Cut-off value was determined by the optimal Youden’s index (sensitivity + specificity − 1). The sensitivity and specificity of the SEPT9 test were calculated as follows: sensitivity = number of true positives/total number of sick individuals in population; specificity = number of true negatives/total number of well individuals in population. 95% CI was calculated according to the efficient-score method (corrected for continuity) (27). P<0.05 was considered statistically significant.
Results
SEPT9 methylation in breast cancer tissues
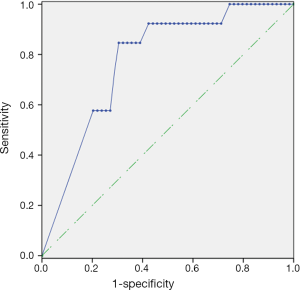
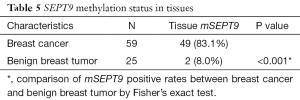
As plotted in Figure 1, ROC curve analysis was performed to evaluate the diagnostic performance of mSEPT9 as a biomarker for differentiating breast cancer from benign breast tumor tissue samples. The calculated AUC was 0.777 and cut-off value of mSEPT9 was 37.6 cycles. As shown in Table 5, mSEPT9 was detected in the tissues of 49 of 59 breast cancer patients (83.1%) and 2 of 25 (8.0%) benign breast tumor patients (two samples were excluded due to the cycle numbers of ACTB >37.6) (P<0.001). The sensitivity and specificity of mSEPT9 for breast cancer detection were 83.1% (95% CI, 71.5–90.5%) and 92.0% (95% CI, 75.0–97.8%), respectively (Table S3).
Full table
Expression of SEPT9_v2 mRNA and SEPT9 protein in tissues
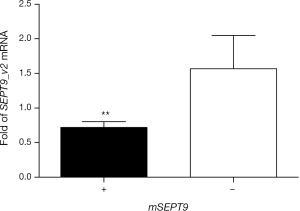
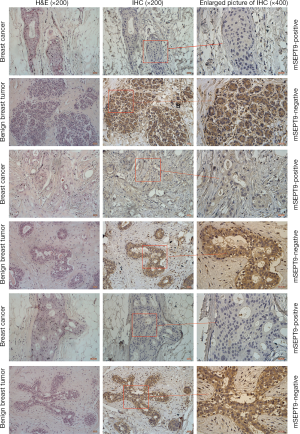
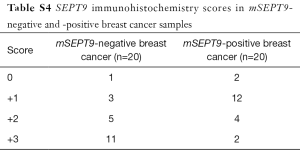
Total RNA was isolated from the biopsy tissues of 40 cases of breast tumors and reverse transcribed. Real-time quantitative PCR assays were carried out to evaluate the levels of SEPT9_v2 mRNA. Expression of SEPT9_v2 mRNA was lower in mSEPT9-positive breast cancer tissues than in mSEPT9-negative benign breast tumor tissues (Figure 2). These biopsy tissues were also analyzed by IHC staining for SEPT9 protein. The SEPT9 was highly expressed in majority of them mSEPT9-negative tumors (IHC grades +2 and +3: 16/20), while either no expressions or low level expressions of SEPT9 were observed in majority of the mSEPT9-positive tumors (0 and +1: 14/20) (Figure 3, Table S4).
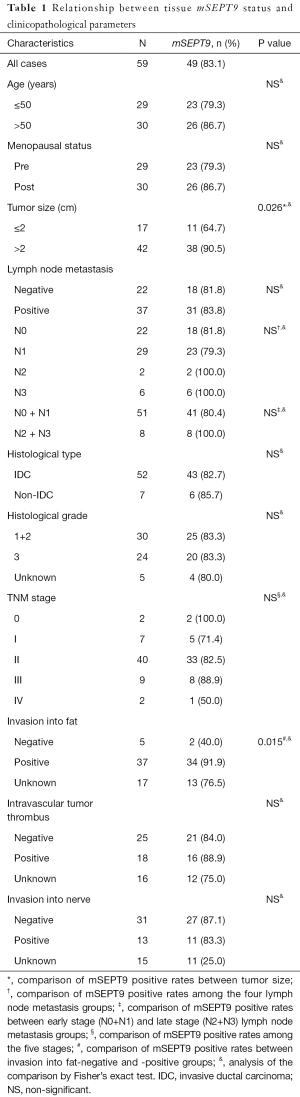
Relationship between tissue mSEPT9 and clinicopathological parameters of breast cancer patients
We investigated the associations between mSEPT9 status in breast cancer tissues and clinicopathological parameters of the breast cancer patients. As presented in Table 1, breast cancer cases with tumor size >2 cm showed a significantly higher positive rate of mSEPT9 than those with tumor size ≤2 cm (90.5% vs. 64.7%, P=0.026). Compared with breast cancer cases with negative fat invasion, the subjects with positive fat invasion showed a higher positive rate of mSEPT9 with statistical significance (91.9% vs. 40.0%, P=0.015). However, no association was found between mSEPT9 and lymph node metastasis, histological type, histological grade, subtype, stage, intravascular tumor thrombus or nerve tissue invasion (P>0.05).
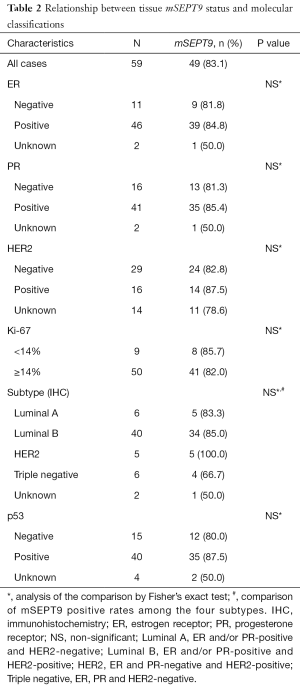
We next sought to examine the associations between mSEPT9 positive rate and the molecular classifications in breast cancer tissues. But unfortunately, no associations between mSEPT9 and ER, PR, HER2, Ki67, or p53 status of the tumors were found (Table 2) (P>0.05).
Methylated SEPT9 genomic DNA in plasma
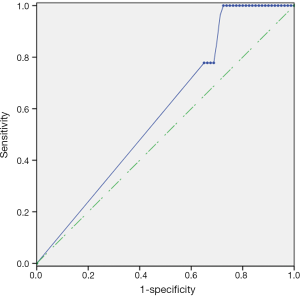
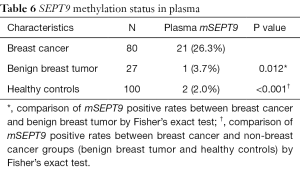
Having researched the status of mSEPT9 in breast cancer tissues, we next examined the methylation status of SEPT9 genomic DNA in the plasma of breast cancer patients. The cut-off value of qPCR for plasma was set at amplification cycles of 40.2, with the pre-requisition of positive ACTB detection before amplification cycles of 32.1 (Figure 4). Plasma mSEPT9 was identified in 21 of 80 (26.3%) breast cancer subjects and 1 of 27 (3.7%) benign breast tumor patients, indicating a significant increase of mSEPT9 in breast cancer patients compared with benign breast tumor patients (P=0.012) (Table 6). As expected, plasma mSEPT9 was detected in only 2 of 100 (2.0%) healthy control subjects, showing a significantly lower positive rate than those in breast cancer cases (P<0.001) (Table 6). The sensitivity and specificity of the plasma mSEPT9 test for breast cancer detection were 26.3% (95% CI, 17.9–36.8%) and 97.6% (95% CI, 93.28–99.19%), respectively (Table S3).
Full table
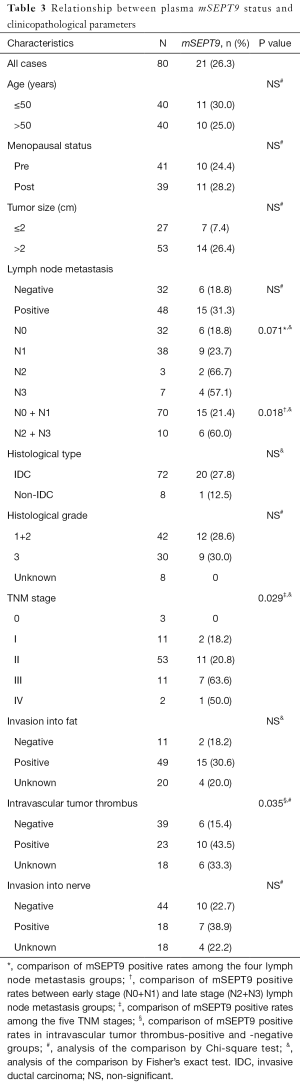
Relationship between plasma mSEPT9 status and clinicopathological parameters of breast cancer
As shown in Table 3, among the 80 patients with breast cancer, the positive rate of plasma mSEPT9 was 0% (0/3) for stage 0, 18.2% (2/11) for stage I, 20.8% (11/53) for stage II, 63.6% (7/11) for stage III, and 50% (1/2) for stage IV in breast cancer patients, suggesting a positive association between mSEPT9 positivity rates and advanced tumor stages (P=0.029). The positive rate of plasma mSEPT9 was 18.8% in stage N0, 23.7% in stage N1, 66.7% in stage N2, and 57.1% in stage N3 breast cancers, respectively, although the differences among the groups were not statistically significant, likely due to the low case numbers of patients with advanced lymph node metastasis. However, when the breast cancer patients were grouped into two lymph node metastasis stage sets [early stage (N0 and N1) and late stage (N2 and N3)], the plasma mSEPT9 positivity rates became significantly different between the two stage sets, with greater plasma mSEPT9 positivity rate in the later stage than in the early stage (P=0.018). Whereas the breast cancer patients with fat or nerve invasion did not show a significant increase in mSEPT9 positivity, patients with intravascular tumor thrombus did show a greater mSEPT9 positive rate than patients without intravascular tumor thrombus (P=0.035).
We also attempted to detect potential associations between plasma mSEPT9 positivity and common molecular classifications of breast cancer. As shown in Table 4, there was a significant difference in the positivity of plasma mSEPT9 between patients with higher Ki67 scores (≥14%) than those with lower Ki67 scores (<14%) (P=0.033). Additionally, the positive rate of plasma mSEPT9 was higher in p53-positive patients than those with p53-negative tumors (P=0.044). However, no associations were found between plasma mSEPT9 positivity and ER, PR and HER2 status.
Comparison of plasma mSEPT9 with CEA, CK19, and CA15-3 test
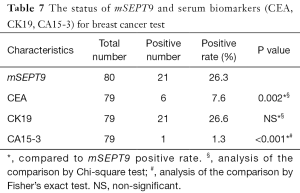
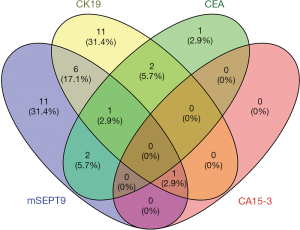
CEA, CK19 and CA15-3 are three most commonly used serum-based breast cancer biomarkers, but they all represent limited sensitivity and specificity (28,29). Coincidentally, for the breast cancer cases already diagnosed by pathological analysis, the positive rates of the CEA, CK19, and CA15-3 tests on plasma samples were 7.6%, 26.6%, and 1.3%, respectively (Table 7). The performances of mSEPT9, CEA, CK19, and CA15-3 test for the diagnosis of breast cancer were shown graphically in Figure 5. These results suggest that there were very limited overlaps between these biomarkers, and mSEPT9 performed comparably with CK19 and better than CEA and CA15-3 in sensitivity.
Full table
Elevated levels of plasma mSEPT9 were decreased in breast cancer patients undergoing anti-cancer treatments
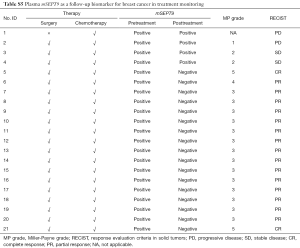
We also analyzed the correlation of plasma mSEPT9 and anti-cancer treatment in 21 patients whose plasma samples were mSEPT9 positive. For this group of patients, plasma specimens were obtained 14 days after the latest treatment. Of the 21 patients, positive to negative conversion was observed in 17 patients (81.0%) (Tables 8,S5). Remarkably, these 17 patients also showed favorable responses to the neoadjuvant chemotherapy, while the remaining 4 patients with persistent plasma mSEPT9 positivity had poor responses after standardized chemotherapy, as verified by the low MP grades (grades 1 and 2; The MP grade is not available for one patient) and persistent or increased tumor sizes.

Full table
Discussion
In the present study, the diagnostic value of mSEPT9 in detecting breast cancer as a noninvasive plasma screening marker for breast cancer was evaluated. We found that mSEPT9 was detected in 83.1% of the breast cancer tissues with a specificity of 92.0%, indicating a remarkable sensitivity as a biomarker for the detection of breast cancer in Chinese women (Tables 5,S3). Because the vast majority of the cancer tissues were mSEPT9 positive, it is not a surprise that sample mSEPT9 positivity was not affected by other clinicopathological parameters or molecular classifications, with exception of tumor size and invasion into fat. As a non-invasive blood breast cancer biomarker, the plasma mSEPT9 assay, in the present setting, still suffers a shortcoming of low sensitivity (26.3%), although the specificity of the mSEPT9 test was 97.6%, which is high for breast cancer detection. Our finding is consistent with a previous study showing a relatively low sensitivity of 11% in patients with breast cancer (15). While the sensitivity of plasma mSEPT9 assay is comparable to CK19 in the detection of breast cancer, the sensitivity is still inadequate as a biomarker for primary screening for breast cancer in women at this stage. A possible explanation for the difference in positivity between tissue and plasma samples is that tissue biopsy sample contains relatively high DNA that is used as template for analysis. Conceivably, early stage breast cancer may release very small amount of DNA in peripheral blood, below the limit of mSEPT9 detection in the current setting, thus making the plasma mSEPT9 assay appear less sensitive. Supporting this speculation is the observation that mSEPT9 positivity was significantly higher in large tumors than in small ones. This is also consistent with the observation that patients with breast cancer of advanced stages, worse lymph node metastasis, intravascular tumor thrombus, higher Ki67 indexes, and p53 protein positivity were more likely to be plasma mSEPT9-positive. It has been shown that in most cancers, the p53 gene is mutated, giving rise to a stable mutant protein whose accumulation is regarded as a hallmark of cancer cells (30). Thus p53 positivity is another indication of more advanced malignant cancer. It is suggested that patients with locally advanced and metastatic cancer are likely to have more tumor cells in circulation leading to more tumor DNA plasma. If lower amounts of ctDNA in the blood are the cause of lower sensitivity of the plasma mSEPT9 test, optimization of the mSEPT9 assay could improve the sensitivity. As our mSEPT9 assay relies on real-time PCR to determine the positivity, relaxing the stringency of the cut-off value by increasing the threshold cycle number of real-time PCR will allow the detection of lower amounts of mSEPT9 DNA. Additionally, lowering the annealing temperature or redesigning of primers covering lower levels of methylation may also significantly increase the sensitivity of the assay methodology. Future studies will focus the fine-tuning of the assay conditions to improve the sensitivity of the mSEPT9 assay for plasma DNA.
In addition to diagnostic value, we found that persistent plasma mSEPT9 positivity after cancer treatment, detected in the current assay setting, was correlated nicely with progressive or stable disease whereas mSEPT9 negative conversion was associated with partial or complete response. It is suggested that the mSEPT9 assay in its present form can already serve as a reliable biomarker for evaluation of the efficacy of the cancer treatments for breast cancer patients with mSEPT9-positive. Moreover, high positive detection rate of plasma mSEPT9 for breast cancer makes mSEPT9 test an ideal modality to distinguish malignancy of tumor at early stages. Of 21 plasma mSEPT9-positive patients before treatments, three patients who completed treatments were found mSEPT9 positive with distant metastases. A possible explanation is that a micro-metastasis was too small to be revealed on imaging study leading to a misjudgment of data interpretation. Given these, use of plasma mSEPT9 as a marker in breast cancer monitoring is advantageous as simplicity and non-radioisotopes.
Despite the intensive quest for a set of noninvasive reliable biomarkers for breast cancer, there appears to have a long way to go before such goal is finally accomplished. While as a biomarker, mSEPT9 is on par with CK19 and far better than CEA and CA15-3 in sensitivity in our hands, a great effort is still required to improve its sensitivity prior to its application as a screening tool for breast cancer in Chinese women. The lower sensitivity and limited overlap between CEA, CK19, and CA15-3 suggest that these biomarkers are unlikely to be useful in cancer screening in the Chinese population.
Our study found that mSEPT9 detection by real-time PCR at the current setting is sensitive to detect breast cancer in biopsy samples. In its current form the mSEPT9 test can be used to evaluate the clinical outcome of cancer treatment for mSEPT9-positive patients. Future studies are needed to improve the sensitivity of the mSEPT9 test to make it a low cost, reliable, and highly sensitive assay for primary screening of breast cancer.

Full table

Full table

Full table
Full table
Full table
Acknowledgments
Funding: This work was supported by National Nature Science Foundation of China (81702884), China Postdoctoral Science Foundation (2017M612290), Nature Science Foundation of Shandong Province (ZR2016HB17) and Medicine and Health Science Technology Foundation of Shandong Province (2015WS0381).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.24). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Liaocheng People
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Wang M, Chen H, Wu K, et al. Evaluation of the prognostic stage in the 8th edition of the American Joint Committee on Cancer in locally advanced breast cancer: An analysis based on SEER 18 database. Breast 2018;37:56-63.
- Klevos GA, Collado-Mesa F, Net JM, et al. Utility of supplemental screening with breast ultrasound in asymptomatic women with dense breast tissue who are not at high risk for breast cancer. Indian J Radiol Imaging 2017;27:52-8. [Crossref] [PubMed]
- Zhang H, Tan H, Gao J, et al. The use of sequential X-ray, CT and MRI in the preoperative evaluation of breast-conserving surgery. Exp Ther Med 2016;12:1275-8. [Crossref] [PubMed]
- Han SH, Yi An Y, Joo Kang B, et al. Takeaways from Pre-Contrast T1 and T2 Breast Magnetic Resonance Imaging in Women with Recently Diagnosed Breast Cancer. Iran J Radiol 2016;13:e36271 [Crossref] [PubMed]
- Wang W, Xu X, Tian B, et al. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta 2017;470:51-5. [Crossref] [PubMed]
- Van De Voorde L, Speeckaert R, Van Gestel D, et al. DNA methylation-based biomarkers in serum of patients with breast cancer. Mutat Res 2012;751:304-25. [Crossref] [PubMed]
- Vo AT, Millis RM. Epigenetics and breast cancers. Obstet Gynecol Int 2012;2012:602720 [PubMed]
- Cheuk IW, Shin VY, Kwong A. Detection of Methylated Circulating DNA as Noninvasive Biomarkers for Breast Cancer Diagnosis. J Breast Cancer 2017;20:12-9. [Crossref] [PubMed]
- Mathot P, Grandin M, Devailly G, et al. DNA methylation signal has a major role in the response of human breast cancer cells to the microenvironment. Oncogenesis 2017;6:e390 [Crossref] [PubMed]
- Perri F, Longo F, Giuliano M, et al. Epigenetic control of gene expression: Potential implications for cancer treatment. Crit Rev Oncol Hematol 2017;111:166-72. [Crossref] [PubMed]
- Wang S, Dorsey TH, Terunuma A, et al. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One 2012;7:e37928 [Crossref] [PubMed]
- Connolly D, Hoang HG, Adler E, et al. Septin 9 amplification and isoform-specific expression in peritumoral and tumor breast tissue. Biol Chem 2014;395:157-67. [Crossref] [PubMed]
- Song L, Li Y. Progress on the clinical application of the SEPT9 gene methylation assay in the past 5 years. Biomark Med 2017;11:415-8. [Crossref] [PubMed]
- Matsui S, Kagara N, Mishima C, et al. Methylation of the SEPT9_v2 promoter as a novel marker for the detection of circulating tumor DNA in breast cancer patients. Oncol Rep 2016;36:2225-35. [Crossref] [PubMed]
- Stanbery L, Petty EM. Steps solidifying a role for SEPT9 in breast cancer suggest that greater strides are needed. Breast Cancer Res 2012;14:101. [Crossref] [PubMed]
- Erratum: Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:345.
- Ogston KN, Miller ID, Payne S, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast 2003;12:320-7. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Huh I, Wu X, Park T, et al. Detecting differential DNA methylation from sequencing of bisulfite converted DNA of diverse species. Brief Bioinform 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Wu D, Zhou G, Jin P, et al. Detection of Colorectal Cancer Using a Simplified SEPT9 Gene Methylation Assay Is a Reliable Method for Opportunistic Screening. J Mol Diagn 2016;18:535-45. [Crossref] [PubMed]
- Hammond ME, Hayes DF, Wolff AC, et al. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract 2010;6:195-7. [Crossref] [PubMed]
- Dowsett M, Nielsen TO, A'Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst 2011;103:1656-64. [Crossref] [PubMed]
- Kobayashi S, Fujimori T, Mitomi H, et al. Immunohistochemical assessment of a unique basal pattern of p53 expression in ulcerative-colitis-associated neoplasia using computer-assisted cytometry. Diagn Pathol 2014;9:99. [Crossref] [PubMed]
- Singh K, Tantravahi U, Lomme MM, et al. Updated 2013 College of American Pathologists/American Society of Clinical Oncology (CAP/ASCO) guideline recommendations for human epidermal growth factor receptor 2 (HER2) fluorescent in situ hybridization (FISH) testing increase HER2 positive and HER2 equivocal breast cancer cases; retrospective study of HER2 FISH results of 836 invasive breast cancers. Breast Cancer Res Treat 2016;157:405-11. [Crossref] [PubMed]
- Seshan VE, Gonen M, Begg CB. Comparing ROC curves derived from regression models. Stat Med 2013;32:1483-93. [Crossref] [PubMed]
- Morey RD, Hoekstra R, Rouder JN, et al. The fallacy of placing confidence in confidence intervals. Psychon Bull Rev 2016;23:103-23. [Crossref] [PubMed]
- Hahn EE, Hays RD, Kahn KL, et al. Use of imaging and biomarker tests for posttreatment care of early-stage breast cancer survivors. Cancer 2013;119:4316-24. [Crossref] [PubMed]
- Henderson MC, Sweeten K, Borman S, et al. dtectDx Breast: A serum biomarker test for breast cancer detection used in conjunction with traditional mammography screening. J Clin Oncol 2013;31:18. [Crossref]
- Rivlin N, Brosh R, Oren M, et al. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011;2:466-74. [Crossref] [PubMed]


