
Overexpressed miR-128a inhibits the proliferation of laryngeal cancer cells
Introduction
Laryngeal squamous cell carcinoma is one of the most malignant tumors of the head and neck, which originates from laryngeal epithelial tissue (1,2). Although significant advances have been achieved in the comprehensive treatment of head and neck cancer, numerous cases remain incurable; thus, it is crucial to pursue novel therapies with improved effects. A profound understanding of the underlying molecular mechanisms of laryngeal carcinoma will assist in the development of better strategies.
MicroRNAs (miRNAs or miRs) are newly recognized, noncoding, regulatory RNA molecules with a length of ~22 nucleotides, have been identified in all Metazoa studied thus far, and it is estimated that the human genome may include 800–1,000 miRNAs (3). Although miRNAs account for a minor fraction of the expressed genome, they are pivotal regulators of development and cellular processes, including cell differentiation, proliferation, morphogenesis, motility, survival and apoptosis (4). The molecules are known to control gene expression through association with the 3'-untranslated region of genes, as well as inhibit protein translation (5). In addition, miRNAs are able to destabilize and mediate the degradation of RNA transcripts (6). They also serve an important role in cancer by controlling the expression levels of certain tumor suppressor genes and oncogenes (7). MiRNA profiling has revealed the distinct expression signatures and important roles of these molecules in various types of human cancer, including glioma (8), lung cancer (9), gastric cancer (10) and prostate cancer (11). Previous studies (12-14) have reported the function of significant miRNAs in head and neck cancer, such as in tongue cancer, oral squamous cell carcinoma and laryngeal squamous cell carcinoma. Furthermore, it has been observed that miR-128 expression was reduced in prostate cancer cells (15), and suppressed prostate cancer by inhibiting BMI-1 and consequently inhibiting tumor-initiating cells (16). Based on our previous research, BMI-1 promotes the progression of laryngeal squamous cell carcinoma (17) and maintains the proliferation of laryngeal cancer stem cells (18), thus suggesting the tumor-suppressive function of miR-128 in laryngeal cancer. However, the role of miR-128 in this carcinoma requires further verification.
In the present study, the aim was to report for the first time the expression of miR-128a in laryngeal squamous cell carcinoma, as well as to investigate whether miR-128a affected the proliferation and apoptosis of laryngeal Hep2 cells following hsa-miRNA-mediated overexpression. By performing a spectrum of functional experiments, the current study provided substantial evidence that miR-128a serves a suppressive role in laryngeal Hep2 cells in vitro and in vivo.
Methods
Primary laryngeal squamous cell carcinoma specimens
In total, 40 pairs of primary laryngeal squamous cell carcinoma samples and adjacent normal tissue were obtained from patients who had provided informed consent, following the resection of laryngeal carcinoma. The tumor samples were collected under the guidance of pathologists and surgeons. All the margins of the specimens were negative, and the tumor samples used in the experiments of the present study were obtained from center of the specimens. All surgical procedures were required to ensure that the tumor margins remained intact. Adjacent normal tissues used as controls were obtained at least 1 cm away from the margins and clinically considered as normal, given the negative margins. All procedures were approved by the Ethics Boards at the Eye and ENT Hospital of Fudan University (Shanghai, China). The clinical and histological characteristics of the patients are presented in Table 1, using the TNM classification system.

Full table
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
All the tissue samples were homogenized in TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and total RNA was extracted according to the protocol suggested by the manufacturer. The total RNA concentration was measured using spectrophotometry at OD260/280. RT was subsequently performed using 1 µg total RNA and M-MLV Reverse Transcriptase (Promega Corp., Madison, WI, USA). PCR amplification was conducted using Taq polymerase (Direct PCR kit, Sangon Biotech Co., Ltd., Shanghai, China), and the miRNA expression was evaluated using the Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). TaqMan probes for the hsa-miR-128a were purchased from Thermo Fisher Scientific, Inc., and RNU6B was used as an endogenous control to standardize the miRNA expression. Thermocycling took place at 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 55 °C for 1 min. All reactions were performed in triplicate. The 2−ΔΔCt formula (19) was used to calculate the miRNA level.
Cell culture
The laryngeal squamous cell carcinoma Hep2 cell line was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cell line was cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a humidified incubator with 5% CO2 at 37 °C.
Vector construction and lentiviral transfection
The transfer vector pLenO-RIP was used to transfer target gene to lentiviruses, and hsa-miR-128 that mimics the mature miR-128a was synthesized. The pre-miRNA sequence was as follows: 5'-UGAGCUGUUGGAUUCGGGGCCGUAGCACUGUCUGAGAGGUUUACAUUUCUCACAGUGAACCGGUCUCUUUUUCAGCUGCUUC-3'. The synthesized pri-miRNA sequence was as follows: 5'-CGACGCGTGATTTTAGGTTTACAAAGCCCTAGCTGTTTTCTGTGTAGCTTTTATTATTCTTATGACTCTTGACAAGTTTGTAGCTTCACCATATACATTTAATATTTTCAATAATTGGCCTTGTTCCTGAGCTGTTGGATTCGGGGCCGTAGCACTGTCTGAGAGGTTTACATTTCTCACAGTGAACCGGTCTCTTTTTCAGCTGCTTCCTGGCTTCTTTTTACTCAGGTTTCCACTGCTTTTTTGCTTTTTTTAATGCTGTATGAAGGTGTTAACATTTGTTTATATTTTTCATTAATTGTAATACCTTTAAATCATGCATCATACTCAGAAATAGGGATTAGGCGGCCGCAA-3'.
NotI and MIuI cloning sites were used for enzyme digestion (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequent to processing enzyme digestion, the lentiviral vector and the target sequences were ligated together. The lentiviral plasmids were then extracted according to the protocol of the AxyPrep Plasmid Miniprep Kit (Axygen; Corning Incorporated, Corning, NY, USA). Next, the hsa-miR-128a lentiviral plasmid was transfected along with packaging plasmids into 293T cells (Gaining Biological, Shanghai, China), and cultured with DMEM with 10% FBS and 1% Glutamax, and then lentiviral titers were measured by flow cytometry following 293T cells transfection. The lentiviruses harboring hsa-miR-128a were then transfected into the laryngeal Hep2 cell line, and the transfection efficiency was evaluated by flow cytometry. Since the transfected cells showing red fluorescence, flow cytometry can recognize the cells and count the percentage of the transfection. Wild-type Hep2 cells served as the control group, and cells transfected with non-targeting plasmids were termed the red fluorescent protein (RFP) group and served as a negative control. Cells transfected with targeting-plasmids were termed the hsa-miR-128a-RFP group. The miR-128a transcription level was confirmed by qPCR.
Cell proliferation assay
The evaluation of the cell proliferation capacity was performed using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) according to the manufacturer’s protocol. Cultured wild-type Hep2 cells, RFP group cells and hsa-miR-128a-RFP group cells were plated in 96-well plates in 0.2 mL RPMI 1640 with 10% FBS at a density of 2×103 cells/well. Each group had six replicates. Proliferation assays were performed each day for 9 days through the addition of 20 µL CCK-8 dye to the wells and incubation for 2 h. The visible light absorbance at 450 nm was measured using a microplate reader (Bio-Rad model 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The cell proliferation capacity was calculated according to the measured optical density (OD), as follows: Cell proliferation (%) = [(test group OD value − culture medium OD value)/(wild-type control group OD value − culture medium OD value)] ×100.
Colony formation assay
Following transfection, the miR-128a overexpression, RFP control and wild-type Hep2 cells were trypsinized with 0.25% trypsin. Cells were seeded into 9.62 cm2 (diameter, 3.5 cm) culture plates at a density of 250 cells per well. Next, 1 mL RPMI 1640 medium with 10% FBS was added to the plates to allow for colony formation and incubated at 37 °C for 7 days. The cultures were then stained with Toluidine blue (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China), and the colony formation rate was calculated as follows: Colony formation (%) = number of colonies/number of transplanted cells.
Apoptosis assay
An apoptosis assay was performed to assess the apoptosis capacity of hsa-miR-128a-RFP and RFP cells. Briefly, cells were centrifuged at 200 ×g for 10 min in 4 °C and suspended in 0.01 M PBS. For flow cytometry analysis, the suspended cells were collected and resuspended in 1× Annexin V Binding Buffer at a concentration of 1×106 cells/mL. A total of 100 µL solution was transferred to a 5 mL culture tube, and stained with 5 µL FITC-Annexin V and 5 µL PI (FITC Annexin V Apoptosis Detection kit I; BD Pharmingen; BD Biosciences, Franklin Lakes, NJ, USA). The solution was cultured for 15 min at room temperature, and then apoptosis was analyzed by a flow cytometry (Beckman Coulter, Inc., Brea, CA, USA). For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) analysis, 2×106 cells/mL cells were suspended in 200 µL PBS and then stained with 50 µL TUNEL reaction mixture (In Situ Cell Death Detection kit, Fluorescein; Roche Diagnostics, Basel, Switzerland). DAPI was used to counterstain the nuclei, and the apoptotic cells were observed under a fluorescence microscope (Leica DM IRB; Leica Microsystems GmbH, Wetzlar, Germany).
In vivo tumorigenicity assay in mice
An in vivo tumorigenicity assay was also performed in the present study using 10 male non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice with an age of 6–8 weeks, which were obtained from Shanghai SLAC Laboratory Animals (Shanghai, China). The NOD/SCID mice were kept in a specific pathogen-free animal center at 24 °C in a humidity of 50%. The weight of the mice ranged from 18–26 g. RFP and hsa-miR-128a cells were suspended in a volume of 200 µL RPMI 1640 medium. Next, the cells were collected and a concentration of 1×105 cells was injected into the subcutaneous space of the axillary fossa of NOD/SCID mice under anesthesia with ketamine/xylazine. A total of 10 mice were injected with RFP cells and 10 were injected with hsa-miR128a cells. During a period of 4 weeks, the mice were examined every 3 days for tumor formation by palpation. Subsequently, all mice were sacrificed by cervical dislocation and tumors were measured in two dimensions with calipers and weighted. The tumor volumes were calculated according to the formula described by Chinnaiyan et al. (20), as follows: Volume = length × width2 × π/6 The experimental protocols were processed according to the guidelines of Shanghai Medical Experimental Animal Care Commission (Shanghai, China).
Statistical analysis
All the results are presented as the mean ± standard deviation of at least three independent experiments. Statistically significant differences were evaluated by independent-samples t-test, analysis of variance and Kruskal-Wallis test. Stata version 12.0 (StataCorp LP, College Station, TX, USA) and Microsoft Office Excel 2003 (Microsoft Corporation, Redmond, WA, USA) were used for data processing and statistical comparison. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
Low expression of miR-128a is observed in laryngeal squamous cell carcinoma
Although miR-128a has been reported to be associated with multiple types of cancer (6-9), it has not been investigated in laryngeal squamous cell carcinoma, to the best of our knowledge. Thus, the present study attempted to determine the association between miR-128a and laryngeal squamous cell carcinoma. Primary tumors were dissected from 40 patients immediately following a laryngectomy. The clinical and histological characteristics of the included patients are presented in Table 1.
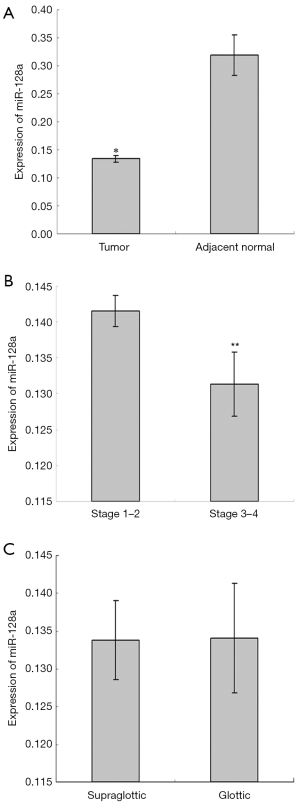
To quantitatively determine the expression of miR-128a in laryngeal squamous cell carcinoma, qPCR was performed on primary tumor specimens and adjacent normal tissues. The results revealed that significantly lower miR-128a expression was observed in laryngeal tumors in comparison with that in normal tissues (Figure 1A). Subsequently, miR-128a expression in different tumor stages and locations was compared. As shown in Figure 1B, the expression of miR-128a was significantly higher in stage 1 and 2 tumor specimens as compared with that in stage 3 and 4 tumors (P<0.05). Regarding the tumor location, there was no evident difference in miR-128a expression between the supraglottic and glottic tumors (Figure 1C).
MiR-128a overexpression by lentiviral plasmids
In order to determine the biological role of miR-128a in laryngeal cancer, a stable hsa-miR-128a overexpression cell line was established. Lentiviruses harboring hsa-miR-128a were constructed and transfected into laryngeal Hep2 cell line, while a non-targeting negative control RFP cell line was also established, with wild-type Hep2 cells considered as the control. All the transfected cells exhibited red fluorescence, and the transfection rate was evaluated by flow cytometry, as shown in Figure 2A. The RFP cells transfected with non-targeting plasmids were observed to have a transfection rate of 80.92%, while the hsa-miR-128a-RFP cells transfected with target plasmids had a transfection rate of 78.84%. The miR-128a transcription level was confirmed by qPCR and the expression of miR-128a in the hsa-miR-128a cells was markedly higher as compared with that in the negative control RFP cells (Figure 2B).

MiR-128a overexpression suppresses cell proliferation in vitro
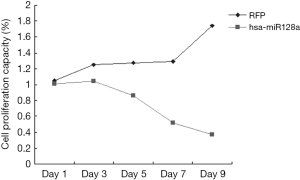
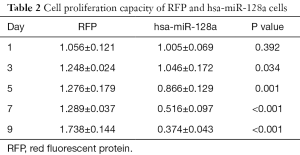
To evaluate the proliferation capacity of miR-128a, Hep2 and hsa-miR-128a cells were cultured in 96-well plates, and the absorbance of the individual cells was measured by a CCK-8 assay on days 1, 3, 5, 7 and 9. As shown in Figure 3 and Table 2, the proliferation capacity of RFP cells was evidently stronger in comparison with that in hsa-miR-128a cells. The negative control cells presented an increasing trend in their proliferation capacity with each day of incubation, while the growth of hsa-miR-128a cells was gradually decreasing. On day 1, there was no significant difference in the proliferation rate between the two groups; however, from day 3, the difference was statistically significant (P<0.05).
Full table
MiR-128a overexpression inhibits laryngeal cancer cell colony formation in vitro
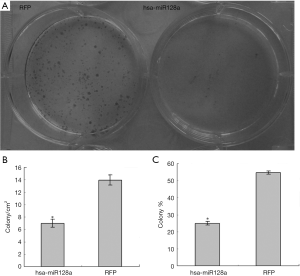
In order to confirm the results of the proliferation assay and to establish a more profound understanding of the effect of miR-128a overexpression, a colony formation assay was performed. Following transfection, hsa-miR-128a and negative control RFP cells were incubated 7 days and then stained with Toluidine blue to examine their colony formation ability. As shown in Figure 4A, increased blue staining of colony spheres was observed in the culture of RFP cells. However, in the culture of hsa-miR-128a cells, colony sphere formation was rare, which was consistent with the results of the proliferation assay. In Table 3, the colony formation capability of the two groups is compared at a quantitative level. The number of colonies formed/cm2 of RFP cells was 13.96±0.83, whereas this value was 7.00±0.67 in the hsa-miR-128a cells, with a significant difference observed (P<0.05; Figure 4B). The percentage of colonies formed compared with the free cells (colony %) further confirmed the results of previous assays. The colony % of RFP cells was 54.66±0.94, while that of hsa-miR-128a cells was 24.98±1.01, revealing a significantly weaker colony formation ability in cells with miR-128a overexpression (P<0.05; Figure 4C).
Full table
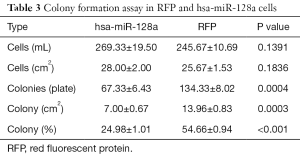
MiR-128a overexpression promotes laryngeal cancer cell apoptosis
As observed in the earlier proliferation assay, miR-128a was capable of inhibiting the growth of laryngeal Hep2 cells. Thus, the study further attempted to examine whether a correlation exists between proliferation and apoptosis in hsa-miR-128a cells by performing apoptosis assays by flow cytometry and TUNEL staining. As observed in the flow cytometry results shown in Figure 5A,B, the percentage of apoptotic cells in the hsa-miR-128a group was 38.73%±0.73%, while the RFP cells demonstrated a significantly reduced apoptosis rate at 4.81%±0.25% (P<0.05). As shown in Figure 5C, the hsa-miR-128a and RFP cells were stained by TUNEL and DAPI, and then observed under a fluorescence microscope. It was observed that hsa-miR-128a cells exhibited enhanced apoptosis as compared with the RFP cells.
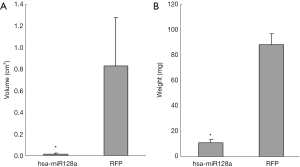
Overexpression of miR-128a inhibits transplanted tumor growth in vivo
To evaluate whether overexpression of miR-128a is able to inhibit the tumorigenic capacity of Hep2 cells in vivo, RFP and hsa-miR-128a cells were injected into the subcutaneous space of the axillary fossa of mice, and tumor formation was examined after 4 weeks. A significant reduction in the tumor volume was observed in the miR-128a overexpression group compared with that in the RFP negative control group (P<0.05), as shown in Figure 6A. The volume of tumors in the RFP group was 0.83±0.45 cm3, while that in the hsa-miR-128a group was 0.015±0.009 cm3. Furthermore, as presented in Figure 6B, a significant decrease in the tumor weight was also confirmed in the hsa-miR-128a group as compared with the RFP group (P<0.05). The tumor weight of the RFP group tumors was 88.00±8.92 mg, while the weight in the hsa-miR-128a group tumors was 10.70±2.75 mg. Therefore, overexpression of miR-128a was able to suppress the tumorigenic ability of Hep2 cells in vivo.
Discussion
As one of the most common and severe malignancies of the head and neck, laryngeal squamous cell carcinoma remains incurable at advanced stages; however, promising comprehensive treatment strategies are being developed. With advances in the understanding of pathogenesis at the DNA level, more profound investigations at the miRNA level should be conducted to obtain novel insight that will assist in the disease treatment. miRNAs have been reported to function as tumor suppressors and oncogenes, and are also referred to as oncomiRs (7). Abnormalities in miRNA expression are considered to serve significant roles in carcinogenesis, which is supported by investigations on the expression of miRNAs in clinical specimens (21,22). For instance, previous studies revealed that let-7 was downregulated in lung tumors (23) as compared with its expression in normal tissue, and miR-127 and miR-128 were downregulated in human bladder cancer (24) and miR-128 in glioma (8), respectively. However, the expression and function of miR-128a remains unclear in laryngeal squamous cell carcinoma. To the best of our knowledge, the present study demonstrated for the first time that miR-128a is downregulated in primary laryngeal tumors, and that overexpression of endogenous miR-128a suppresses the cell proliferation and promotes apoptosis. Therefore, targeting miR-128a may be a novel method to promote the treatment of laryngeal squamous cell carcinoma.
In the present study, 40 primary laryngeal squamous cell carcinoma specimens along with 40 adjacent normal epithelial tissue samples were collected. The majority of the included patients were males with an age of >50 years, with an advanced stage of laryngeal cancer and tumors mainly located in the supraglottic region, without lymph node metastasis. qPCR indicated that miR-128a exhibited a significantly lower expression in the tumor samples as compared with the adjacent normal tissue, which is consistent with previous findings on the expression of this miRNA in glioma tissues by Ciafrè et al. (8) and Zhang et al. (25). However, according to the study by Lu et al. (26), miR-128a was upregulated in oral cancer cell lines, which may be caused by the primary tumor location difference. Further research on the signal pathways of the miR-128a is required to better understand this observation. The current study also observed that the expression of miR-128a in stage 1 and 2 tumors was significantly higher in comparison with that in stage 3 and 4 tumors; however, there was no statistically significant variation between the supraglottic and glottic tumors. These results indicated that the expression of miR-128a may be associated with the survival of the laryngeal cancer patients, although further investigations are required to confirm this.
A cell proliferation assay was also performed in the current study to investigate whether the overexpression of miR-128a affected the growth of Hep2 cells in vitro. Using the CCK-8 method, it was observed that the proliferation of hsa-miR-128a cells presented a declining trend, while the proliferation of negative control RFP cells appeared to increase each day. This result indicated that hsa-miR-128a transfection not only inhibited the proliferation of Hep2 cells, but also promoted the apoptosis. In the study by Godlewski et al. (27), it was observed that a short hairpin oligonucleotide mimic of pre-miR-128-1 led to high levels of mature miR-128 expression, and significantly reduced the proliferation of U87 and U251 glioma cell lines as compared with a control oligonucleotide. Furthermore, other previous studies (25,28) revealed that miR-128 presented the capacity to suppress the proliferation of cancer cells. The findings of the present study are consistent with the aforementioned previous observations, in particular regarding the cell growth. In order to confirm the results of the proliferation assay, a colony formation assay was also performed in the current study. The colony/cm2 and colony % of the hsa-miR-128a cells were markedly reduced in comparison with the RFP cells, which also confirmed the observations of the proliferation assay. Apoptosis assays, including Annexin V and TUNEL staining, were subsequently conducted to investigate whether hsa-miR-128a was able to promote apoptosis. It was demonstrated that the apoptosis rate of hsa-miR-128a cells was almost 10-fold greater compared with that of the RFP cells, which indicated that miR-128a not only affected the pathway of proliferation and apoptosis, but also that there may be a crosslink between the two pathways. These findings are novel, and no relevant previous reports exist on this function of miR-128a. In subsequent studies, the signaling pathways of proliferation and apoptosis will be further investigated.
In order to prove the function of miR-128a on the proliferation of laryngeal cancer cells, a tumorigenicity assay was also conducted in the present study. A total of 10 male NOD/SCID mice (6–8 weeks old) were injected with RFP and miR-128a overexpression cells at the subcutaneous space of their axillary fossa under anesthesia. Mice were examined every 3 days for tumor formation by palpation for 4 weeks after injection. After sacrifice, the tumor volume of the RFP group tumors was markedly larger in comparison with that of the hsa-miR-128a group tumors. It was also revealed that the weight of tumors in the hsa-miR-128a group was significantly lower compared with that in the RFP group. According to the study by Jin et al. (16), miR-128 overexpression inhibited Du145 tumor growth and reduced the tumor incidence rate, providing the first in vivo evidence that miR-128 possesses prostate cancer-suppressive effects. The present study results were consistent with these previous observations, further suggesting that miR-128a suppressed the tumor growth of laryngeal squamous cell carcinoma.
In conclusion, the current study reported for the first time that miR-128a is downregulated in laryngeal squamous cell carcinoma. In addition, it was demonstrated that overexpression of miR-128a inhibited the proliferation capacity and promoted the apoptosis of laryngeal Hep2 cells. Furthermore, upregulation of miR-128a suppressed the tumor formation in vivo. Taken together, these results suggest that upregulation of miR-128a is a novel possible target for laryngeal cancer treatment.
Acknowledgments
The authors would like to thank Ms. Yang Wang, Fudan University, Shanghai, China for her great help in the molecular biology experiments.
Funding: The present study was supported by the National Natural Science Foundation of China (grant no. 81402237), and the Natural Science Foundation of Anhui Province, China (grant no. 11040606M207).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.06.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from the patients. All procedures were approved by the Ethics Boards at the Eye and ENT Hospital of Fudan University (Shanghai, China).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mastronikolis NS, Tsiambas E, Papadas TA, et al. Deregulation of PTEN expression in laryngeal squamous cell carcinoma based on tissue microarray digital analysis. Anticancer Res 2017;37:5521-4. [PubMed]
- Ma Z, Zhang H, Lian M, et al. SLC7A11, a component of cysteine/glutamate transporter, is a novel biomarker for the diagnosis and prognosis in laryngeal squamous cell carcinoma. Oncol Rep 2017;38:3019-29. [Crossref] [PubMed]
- Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genet 2005;37:766-70. [Crossref] [PubMed]
- Liu X, Chen Z, Yu J, et al. MicroRNA profiling and head and neck cancer. Comp Funct Genomics 2009;837514 [PubMed]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development 2005;132:4645-52. [Crossref] [PubMed]
- Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science 2005;308:833-8. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 2005;334:1351-8. [Crossref] [PubMed]
- Wang N, Liang H, Zhou Y, et al. miR-203 suppresses the proliferation and migration and promotes the apoptosis of lung cancer cells by targeting SRC. PLoS One 2014;9:e105570 [Crossref] [PubMed]
- Shen J, Niu W, Zhou M, et al. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PLoS One 2014;9:e104510 [Crossref] [PubMed]
- Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol 2015;67:33-41. [Crossref] [PubMed]
- Lin Z, Sun L, Chen W, et al. MiR-639 regulates transforming growth factor beta-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting FOXC1. Cancer Sci 2014;105:1288-98. [Crossref] [PubMed]
- Hedbäck N, Jensen DH, Specht L, et al. MiR-21 expression in the tumor stroma of oral squamous cell carcinoma: an independent biomarker of disease free survival. PLoS One 2014;9:e95193 [Crossref] [PubMed]
- Tian L, Li M, Ge J, et al. MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumors. Tumour Biol 2014;35:5953-63. [Crossref] [PubMed]
- Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 2008;68:6162-70. [Crossref] [PubMed]
- Jin M, Zhang T, Liu C, et al. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res 2014;74:4183-95. [Crossref] [PubMed]
- Chen H, Zhou L, Wan G, et al. BMI1 promotes the progression of laryngeal squamous cell carcinoma. Oral Oncol 2011;47:472-81. [Crossref] [PubMed]
- Chen H, Zhou L, Dou T, et al. BMI1's maintenance of the proliferative capacity of laryngeal cancer stem cells. Head Neck 2011;33:1115-25. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Chinnaiyan AM, Prasad U, Shankar S, et al. Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci U S A 2000;97:1754-9. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005;353:1793-801. [Crossref] [PubMed]
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [Crossref] [PubMed]
- Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006;9:435-43. [Crossref] [PubMed]
- Zhang Y, Chao T, Li R, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl) 2009;87:43-51. [Crossref] [PubMed]
- Lu YC, Chen YJ, Wang HM, et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profling. Cancer Prev Res (Phila) 2012;5:665-74. [Crossref] [PubMed]
- Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 2008;68:9125-9130. [Crossref] [PubMed]
- Venkataraman S, Alimova I, Fan R, et al. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One 2010;5:e10748 [Crossref] [PubMed]


