
The TGF-β-regulated X-inactive specific transcript inhibits papillary thyroid cancer migration and invasion
Introduction
Papillary thyroid cancer (PTC), a subtype of the most common endocrine gland malignancy, is involved in 85% of thyroid malignancy patients. PTC can progress from multiple thyroid diseases, such as goiter, thyroiditis, or hyperthyroidism (1). Because of its slow development and relatively good prognosis, PTC has seldom been studied (2,3). Lymph node metastasis does occur in PTC patients, and according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, the prognosis of PTC patients will become worse as soon as the lymph nodes are invaded (4,5). Additionally, there is a group of patients with highly malignant PTC, which expresses a higher growth rate and a significantly higher probability of lymph nodes metastasis, even distant metastasis. In addition, the resection range of lymph node metastasis patients is usually larger than lymph node negative patients, which is related to the quality of life for postoperative patients. Thus, it is important to determine the underlying mechanisms of PTC metastasis, and thereby improve prognoses and treatments.
Long non-coding RNAs (lncRNAs) comprise a class of transcripts longer than 200 nucleotides, which lack the function of protein coding. Studies have shown that lncRNAs play a crucial role in cancer initiation, progression, and metastasis (6,7). Thousands of lncRNAs can be screened in a human transcriptome, but only a few have been well-studied in terms of thyroid cancer, such as PTCSC2 (8), H19 (9) and NEAT1 (10). LncRNA X-inactive specific transcript (XIST), a key player in X-chromosome inactivation that encodes an RNA that codes the silent X chromosome, has been found to be associated with multiple malignancies, such as breast cancer, lung cancer, and gastric cancer (11-13). However, its functions in PTC have not been reported.
Through bioinformatics analyses using available databases, we found that lncRNA XIST expression was significantly higher in PTC tissues without lymph metastasis than that in metastatic PTC tissues. Moreover, there was a correlation between XIST expression profiles and TGF-β, a signaling pathway which is associated with cancer cell proliferation and metastasis (14). We then determined the functions of lncRNA XIST and its association with the TGF-β pathway components in PTC cells.
Methods
Bioinformatic analysis
Transcriptome expression profile of thyroid cancer patients and their clinical data were gathered from the TCGA data sets (TCGA-THCA-exp-HiSeqV2-PANCAN-2015-02-24, n=286) and analyzed with R software (“limma”). We selected the PTC patients including 62 patients with lymph nodes metastasis and 77 lymph nodes negative ones and compared their different RNAs. Gene ontology (GO) enrichment was analyzed with DAVID (https://david.ncifcrf.gov). GO enrichment was classed with adjusted p values, and top 10 were accepted. Key molecules were analyzed with KEGG PATHWAY Database (http://www.kegg.jp). Pearson correlation analysis was performed with R (“cor”) and the correlation coefficient was shown with absolute value. In order to observe the results easily, the hot map color of positive and negative correlation was unified.
Study population
A total of 60 cases of PTC tissues were collected from the patients who were diagnosed with PTC and underwent surgical resection between Sep 2016 and May 2017 at the Department of Thyroid Surgery, Affiliated Hospital to Qingdao University. 30 of them were diagnosed with lymph nodes metastasis, and the other 30 without. All patients did not undergo preoperative radiotherapy or chemotherapy, since those who had received preoperative radiotherapy or chemotherapy and those with recurrence or distant metastasis were excluded. Additionally, the paraffin-embedded tissues were collected from 40 of the PTC patients above in the Pathology Department, Affiliated Hospital to Qingdao University. All the individuals involved in this study were signed informed consent and approved by the ethics committee of Affiliated Hospital of Qingdao University (QDFYLLW201605 NO.05-67). About 0.3-mm thick slices were cut, dried, deparaffinized, and rehydrated for in situ hybridization (ISH) and immunohistochemistry (IHC) assays.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the tissues and cells cultured with TRIzol reagent (Solarbio, China). The SuperScript® III First-Strand Synthesis System for RT-PCR (Thermo Fisher Scientific, USA) was used to reversely transcribe RNA into cDNA. QRT-PCR was performed on the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, USA) with SYBR® Green PCR Master Mix (Thermo Fisher Scientific, USA). The primers were as follow: XIST, forward 5'-GGTTCTCTCTCCAGAAGCTAGGAAAG, reverse 5'-TGGTAGATGGCATTGT GTATTATATGG; TGF-β forward, 5'-AACCCACAACGAAATCTA, reverse, 5'-TGAGGTATCGCCAGGAAT; Smad2 forward 5'-GATCCCACCAGGCTGTAACC, reverse 5'-GCCTCGATATTCTGCTCCC; Smad3 forward 5'-CCAGCACACAATAACTTGGA, reverse 5'-AGACACACTGGAACAGCGGA; Smad4 forward 5'-CAGCTATGCCAGAAGCCAGA, reverse 5'-GAACTCCTGGGACTTTCAACTGAC. GAPDH was used for the internal control of forward 5'-CCATGGAGAAGGCTGGGG, reverse 5'-CAAAGTTGTCAT GGATGACC.
Relative RNA levels were calculated with 2−ΔCt [where ΔCt = Ct(gene) − Ct(reference gene)]. Additionally, the fold change (FC) of RNA expression was calculated with the 2−ΔΔCt method. PCR for each sample was performed three times.
ISH and IHC
The probe of ISH for XIST RNA expression analysis was published by HaoGe Biological Technology Co (Shanghai, China). In brief, the paraffin-embedded TMAs were incubated with Digoxigenin-labeled DNA probes complementary to XIST RNA. After detection of Digoxigenin-labeled Zyto-Fast chromogenic ISH (CISH) probes and hybridization, incubation was performed with a secondary antibody for visualization. The IHC procedure was conducted with the avidin-biotin-peroxidase complex (ABC) method. The antibody used in the experiment included goat anti-TGF-β1 polyclonal antibody (1:500, ab92486, Abcam, USA) and anti-Smad4 (1:500, ab40759, Abcam, USA). Images were acquired using OLYMPUS BX43 microscope (Olympus Corporation, Tokyo, Japan). Five vision fields were randomly chosen in each pathological section. The expression rate was calculated with ImagePro Plus software (MEDIA CYBERNETICS, USA). Negative control tests were done via primary antisera pre-absorption using its respective antigen.
Cell culture
The human thyroid papillary cancer cell lines TPC-1 and BCPAP were obtained from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, Massachusetts) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Thermo Fisher Scientific), 100 U/mL penicillin, and 100 mg/mL streptomycin (Thermo Fisher Scientific). Next, they were cultured in an incubator with 5% CO2 at 37 °C.
Plasmid conduction and transfection
The XIST and TGF-β sequence was synthesized and subcloned into the pcDNA3.1 (Genechem, Shanghai, China) vector. Eukaryotic Expression Plasmid of shRNA of XIST and TGF-β mRNA were also constructed (Genechem, Shanghai, China). PTC and BCPAP cells were transfected for 7 hours, and then the transfection media was replaced with the normal media which were used for transfection for another 24 hours. Then, the transfected cells were chosen in the medium containing puromycin. The plasmid which has higher transfection efficiency was selected for further assays. Each experiment was performed three times.
Cell migration and invasion assay
Transwell chamber was employed to detect cell migration and invasion ability (8.0-µm pore size polycarbonate filters; BD Biosciences). Briefly speaking, after 48 hours after transfection, the cells (2×105 cells/well) were resuspended in 200 µL serum-free RPMI 1640 medium and seeded on the upper compartments of the chamber, and then, 600 µL complete medium was added into the lower compartments. In the invasion assay, matrix glue (Qcbio, Shanghai, China) was added onto the upper surface of the polycarbonate membrane filter. The cells were incubated at 37 °C for 48 hours before non-traversed cells were removed with a cotton swab; 100% methanol (Thermo Fisher Scientific) was used to fix cells on the lower side of the filter, and then crystal violet was used to stain cells. We counted the cells with microscope (Leica Microsystems, Wetzlar, Germany) at a 100 magnification. Five visual fields were randomly chosen in each well.
Western blot
The total proteins of cells or tissues were extracted with total protein extraction kit (Jiancheng Bioengineering Institute, Nanjing, China). And then, anti-TGF-β1 polyclonal antibody (1:1,000, ab92486, Abcam, Cambridge, UK), anti-Smad4 (1:1,000, ab40759, Abcam, Cambridge, UK), and anti-Smad2/3 (1:1,500, ab63399, Abcam, Cambridge, UK) were used as the primary antibody. β-actin antibody (No. ab8227; 1:1,500 diluted; Abcam, Cambridge, UK) was used as a control. Goat anti-rabbit secondary antibody IgG (HRP) (No. ab7090; 1:5,000 diluted; Abcam, Cambridge, UK) was used to incubate for 1 hour.
Statistical analyses
All data were analyzed with SPSS 23.0 (SPSS Inc., Chicago, IL, USA), Microsoft Excel (Microsoft, Washington, USA) and GraphPad Prism (GraphPad Software, La Jolla, California, USA). Microsoft PowerPoint (Microsoft, Washington, USA) was used to modify the figure size. The data were represented as mean ± SD. Chi square test and t-test were used to determine the significant differences between the two groups, and P<0.05 were considered to be statistically significant (*P<0.05, **P<0.01, ***P<0.005, ****P<0.001).
Results
Bioinformatics analyses indicate that lncRNA XIST is associated with lymph node metastasis of PTC
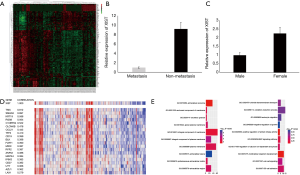
In order to study the differences of the transcriptomes between lymph node metastasis PTC and non-metastasis PTC, we performed a microarray analysis using a thyroid cancer transcriptome from the TCGA database, and then performed a series of in silico hybridization analyses between PTC patients with and without lymph node metastasis [N1 + N2 (n=77) vs. N0 (n=62)]. Eventually, we identified 198 differential RNAs (logFC >2) (Figure 1A). Among the 198 differential RNAs, we found that lncRNA XIST, which was downregulated in the lymph node metastasis cohort, showed the most significant difference (metastasis vs. non-metastasis, FC =9.28; P=1.18×10−6) (Figure 1B; Table S1). According to previous studies, XIST is links to X chromosome and its expression is related to female individuals. Then, we analyzed XIST expression in different sex. The result indicated that XIST is higher in female PTC patients than males (Figure 1C) (FC =2.26; P=0.0002; females, n=68; males, n=71). Next, Pearson’s correlation analyses were performed to identify which of these 198 molecules were closely related to XIST. According to the results, we found that there was a significant negative correlation between XIST and Smad4, a core protein in the TGF-β superfamily (correlation coefficient =0.697) (Figure 1D). In contrast to the downregulation of XIST, Smad4 expression was elevated in lymph node metastasis PTC (metastasis vs. non-metastasis, logFC =−2.60; P=4.16×10−5) (Table S1). Furthermore, A GO enrichment analysis was also performed. From the results, cell component (Figure 1E, left) and biological process (right) were identified as relevant. Interestingly, the BMP pathway, a signaling pathway closely related to the TGF-β pathway (15), was identified in the GO analyses (Figure 1E, right). These analyses inferred that XIST was associated with PTC metastasis, and may also have some interactions with Smad4, or with the TGF-β signaling pathway.
TGF-β and Smad4 proteins are increased and XIST is decreased in lymph node metastatic PTC patients
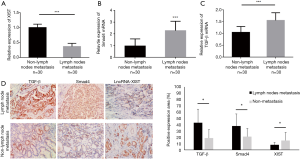
To further determine the role of lncRNA XIST in PTC, we collected 60 cases of PTC tissue samples during operations, and then detected the expression profiles of lncRNA XIST. After comparing the expression profiles of XIST between the lymph node metastasis PTC patients (Cohort 1) and non-metastasis patients (Cohort 2), we found that this RNA was significantly lower in Cohort 1 than in Cohort 2 (FC =−2.83; P=0.0037), which was in agreement with the in silico analyses discussed above (Figure 2A). In order to confirm the bioinformatics results, we also detected the mRNA levels of Smad4 and TGF-β in the two cohorts. The results indicated that mRNA [FC =2.37; P=0.0047 (Figure 2B), and FC =1.61; P=0.0039 (Figure 2C)] of Smad4 and TGF-β, respectively, were elevated in lymph node metastasis PTC patients. The results showed that XIST was decreased while TGF-β and SMAD4 mRNA levels were elevated in lymph node metastasis PTC patients when compared with those of non-metastasis PTC patients. The samples from the 60 cases of PTC were paraffin embedded, and the protein expressions of Smad4 and TGF-β, and the RNA expression of XIST were detected using IHC and ISH methods, respectively. The results showed that all three molecules were mainly localized in the nucleus, indicating that there was a possibility of interactions among them in PTC cells (Figure 2D, left). In addition, in most samples from patients with lymph node metastasis, TGF-β and Smad4 expressions were higher, while XIST expression was lower than in non-metastasis patient samples (Figure 2D, right; P<0.05). These results demonstrated that TGF-β and Smad4 were significantly increased and XIST was decreased in lymph node metastasis PTC patients.
XIST functions as an inhibitor of PTC cell invasion and migration in vitro
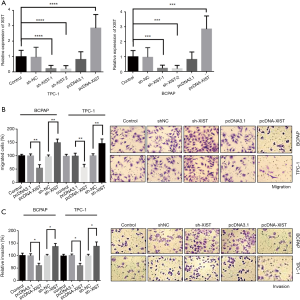
To study the association between XIST and PTC cell invasion and migration abilities, we performed cell migration and invasion assays in two PTC cell lines, BCPAP and TPC-1. Transfections were performed in both BCPAP and TPC-1 cells to detect overexpression or knockdown of XIST, respectively. The transfection efficiency of overexpressed plasmids and two shRNA plasmids were detected in both cell lines, suggesting that XIST was effectively overexpressed or knocked down (Figure 3A; P<0.005). Thereafter, Transwell® migration and invasion assays were performed to detect the effects of XIST in PTC cells. Cell migration assays revealed that XIST overexpression decreased the cell migration by 50% and 48% in BCPAP and TPC-1 cells, respectively, while XIST knockdown increased the cell migration by 51% and 46% in the two cell lines, respectively (Figure 3B, left, and images at right). Cell invasion assays revealed that cell invasion was decreased by 44% and 47%, and by 36% and 38% with XIST overexpression and XIST knockdown, respectively (Figure 3C, left, and images at right). The results suggested that XIST functions as an inhibitor of PTC cell invasion and migration in vitro.
TGF-β functions as an inhibitor of XIST expression in vitro
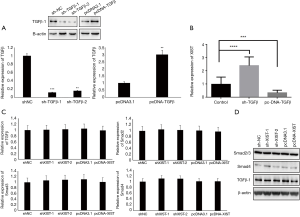
Because there were some correlations between XIST and TGF-β/Smad4 in PTC patients, we performed assays to further study the relationship between XIST and TGF-β, as well as several core members of the TGF-β family. First, TGF-β was overexpressed and knocked down in TPC-1 cells, and the mRNA and protein levels were detected after transfections (Figure 4A; P<0.01). We then detected the changes of XIST expression in different TGF-β-transfected TPC-1 cells. XIST expression was significantly upregulated in TGF-β knockdown cells (FC =2.3; P<0.001), while it was downregulated in TGF-β overexpression cells (FC =−0.32; P<0.005) (Figure 4B). Next, we performed assays to confirm the functions of XIST in the TGF-β pathway. We chose several core proteins in the TGF-β pathway, including Smad2, Smad3, Smad4, and TGF-β1, and detected mRNA expression levels and protein levels in different XIST-transfected cells using qRT-PCR and western blotting, respectively. While the results of qRT-PCR and western blotting demonstrated that there were no changes of Smad2, Smad3, Smad4, and TGF-β1 in either protein or mRNA levels (all P>0.05) (Figure 4C,D). These results indicated that TGF-β functions to inhibit XIST expression, while XIST does not regulate the TGF-β pathway in PTC cells.
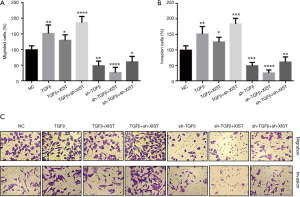
TGF-β inhibits XIST expression and thereby promotes PTC cell invasion and migration
Although TGF-β was shown to function as an inhibitor of XIST in PTC cells, it was still unclear if the interactions between TGF-β and XIST regulated PTC cell invasion and migration. In order to study how the interactions between TGF-β and XIST regulated PTC cell invasion and migration, we performed co-transfection assays and Transwell® invasion and migration assays in TPC-1 cells (Figure 5). The results demonstrated that overexpression of TGF-β improved both cell migration and invasion by 47% and 50%, respectively. We have shown that XIST was downregulated in TGF-β-overexpressing cells; therefore, we overexpressed XIST in TGF-β-overexpressing cells and observed the changes in cell migration and invasion abilities. The results showed that cell migration and invasion abilities were decreased in the TGF-β-overexpressing cells (125% and 126%, respectively). We then knocked down XIST in the TGF-β-overexpressing cells, and cell migration and invasion abilities were significantly increased (186% and 179%, respectively). In addition, Transwell® invasion and migration assays showed that cell migration and invasion were significantly suppressed (52% and 47%, respectively) after knockdown of TGF-β in TPC-1 cells. Increased expression levels of XIST in the TGF-β knockdown cells inhibited the migration and invasion (32% and 25%, respectively). Finally, we decreased XIST in TGF-β knockdown cells, and found that migration and invasion were elevated (68% and 62%, respectively). These results showed that TGF-β promotes migration and invasion via inhibition of XIST expression in PTC cells.
Discussion
In this study, we demonstrated that lncRNA XIST functioned as an inhibitor of cancer cell metastasis in PTC patients and in PTC cells in vitro. We also found that TGF-β suppressed XIST expression in PTC cells, thereby regulating cell metastasis. The regulation mechanisms of XIST expression have been reported in previous studies. Sripathy et al. (16) reported that in Rett syndrome, the activity of the core molecule on the X chromosome, MeCP2, was regulated by BMP/TGF-β superfamily components. They further showed that overexpression of BMP promoted XIST expression, while overexpression of TGF-β decreased XIST expression. These results were in agreement with those of our study. In other studies, it was revealed that TSIX, the antisense RNA of XIST, inhibited XIST expression. It was also reported that XIST and TSIX were co-transcribed RNAs, and their transcription dynamics was regulated by the ratio of X chromosome and autosome, as well as by the semi-stable transcriptional states (17,18). Our results also showed close association between XIST and TXIS (Figure 1C), which indicates that XIST expression is also regulated by TSIX in PTC.
The role of XIST has already been explored in previous studies. XIST has been shown to regulate the spreading and silencing of chromatin in female mammals (19). In breast cancer, XIST is also mainly associated with the coating and genetic instability of the X chromosome. After being localized to the X chromosome, XIST recruits multiple factors indirectly to execute X chromosome inactivation, thereby regulating cancer cell activities (13,20,21). McHugh et al. reported that XIST silences transcription by directly interacting with SHARP, recruiting SMRT, activating HDAC3, and deacetylating histones to exclude Pol II across the X chromosome (22). Moreover, in vivo assay results indicated that XIST RNA is a potent suppressor of hematologic cancer. XIST loss results in X reactivation and consequent genome-wide changes, thereby leading to cancer (23). XIST was also studied as a biomarker of breast cancer treatment effects. The study found that histone deacetylase inhibitor, Abexinostat, can be used as a differentiation therapy, targeting the breast cancer stem cells, and this therapy causes a significant decrease in XIST expression (24). These studies suggest that the main function of XIST in the nucleus is to bind to the X chromosome, and then, coordinate the expression and silencing of different genes. The underlying mechanism, that XIST inhibits PTC cell migration and invasion, may be associated with some metastasis-related genes that are overexpressed or silenced by XIST. According to previous studies, XIST is links to X chromosome and its expression is related to female individuals. Our result showed that XIST was higher in female PTC patients than males. That is coincident with other studies. This also indicated that our conclusion is more accurate in female people. Some studies have also reported that lncRNA XIST functions as a regulator of the TGF-β pathway or as an oncogene by silencing tumor suppressor genes in lung cancer or other malignancies, which is different from our results (11,25). These differences in results could be due to the known cell type- and context-specific effects of the TGF-β superfamily. Thus, in different cell microenvironments, the biological mechanisms can be changed, and XIST may regulate different gene expressions.
In this study, we analyzed the mechanisms of PTC lymph node metastasis, and found that lncRNA XIST exerts important functions in this process. We studied XIST and its correlated proteins and confirmed the inhibition function of TGF-β against this lncRNA in PTC cells. Our study provides a new biomarker of PTC patient metastasis and prognosis. For postoperative patients, XIST expression levels can be detected using tumor tissue biopsies. Patients with elevated XIST in tumor samples may indicate an increased metastasis possibility, to which more attention should be paid in the clinic. According to some studies reporting that XIST can be a biomarker of breast cancer treatment effects (24), XIST may also have a potential to serve as a therapeutic target in PTC prevention and treatment. In future studies, we will focus on the detailed mechanisms of the interactions between TGF-β and XIST in PTC. More studies are also necessary to reveal the molecular mechanisms of XIST inhibition of PTC metastasis.
Conclusions
XIST was regulated by TGF-β, functioned as an inhibitor of PTC metastasis, and therefore provides a new biomarker of PTC patient metastasis and prognosis.
Full table
Acknowledgments
Writing support was provided by International Science Editing.
Funding: This work was supported by the National Natural Science Foundation of China (81571625).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.07.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Affiliated Hospital of Qingdao University (QDFYLLW201605 N0.05-67) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fahiminiya S, de Kock L, Foulkes WD. Biologic and Clinical Perspectives on Thyroid Cancer. N Engl J Med 2016;375:2306-7. [Crossref] [PubMed]
- Randle RW, Bushman NM, Orne J, et al. Papillary Thyroid Cancer: The Good and Bad of the "Good Cancer". Thyroid 2017;27:902-7. [Crossref] [PubMed]
- Rusinek D, Chmielik E, Krajewska J, et al. Current Advances in Thyroid Cancer Management. Are We Ready for the Epidemic Rise of Diagnoses? Int J Mol Sci 2017;18. [PubMed]
- Lamartina L, Grani G, Arvat E, et al. 8th edition of the AJCC/TNM staging system of thyroid cancer: what to expect (ITCO#2). Endocr Relat Cancer 2018;25:L7-L11.
- Tuttle RM, Haugen B, Perrier ND. Updated American Joint Committee on Cancer/Tumor-Node-Metastasis Staging System for Differentiated and Anaplastic Thyroid Cancer (Eighth Edition): What Changed and Why? Thyroid 2017;27:751-6.
- Huarte M. The emerging role of lncRNAs in cancer. Nature medicine 2015;21:1253-61. [Crossref] [PubMed]
- Cui M, You L, Ren X, et al. Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun 2016;471:10-4. [Crossref] [PubMed]
- Wang Y, He H, Li W, et al. MYH9 binds to lncRNA gene PTCSC2 and regulates FOXE1 in the 9q22 thyroid cancer risk locus. Proc Natl Acad Sci U S A 2017;114:474-9. [Crossref] [PubMed]
- Wang P, Liu G, Xu W, et al. Long Non-coding RNA H19 Inhibits Cell Viability, Migration, and Invasion Via Downregulation of IRS-1 in Thyroid Cancer Cells. Technol Cancer Res Treat 2017;16:1102-12. [Crossref] [PubMed]
- Zhang H, Cai Y, Zheng L, et al. Long non-coding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J Cell Physiol 2018;
- Li C, Wan L, Liu Z, et al. Long non-coding RNA XIST promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett 2018;418:185-95. [Crossref] [PubMed]
- Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res 2016;35:142. [Crossref] [PubMed]
- Xiao C, Sharp JA, Kawahara M, et al. The XIST non-coding RNA functions independently of BRCA1 in X inactivation. Cell 2007;128:977-89. [Crossref] [PubMed]
- Alderton G. Tumour microenvironment: To me, to you. Nat Rev Cancer 2013;13:756-7. [Crossref] [PubMed]
- Ohhata T, Senner CE, Hemberger M, Wutz A. Lineage-specific function of the non-coding Tsix RNA for Xist repression and Xi reactivation in mice. Genes Dev 2011;25:1702-15. [Crossref] [PubMed]
- Sripathy S, Leko V, Adrianse RL, et al. Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-beta superfamily as a regulator of XIST expression. Proc Natl Acad Sci U S A 2017;114:1619-24. [Crossref] [PubMed]
- Deuve JL, Bonnet-Garnier A, Beaujean N, et al. Antagonist Xist and Tsix co-transcription during mouse oogenesis and maternal Xist expression during pre-implantation development calls into question the nature of the maternal imprint on the X chromosome. Epigenetics 2015;10:931-42. [Crossref] [PubMed]
- Loos F, Maduro C, Loda A, et al. Xist and Tsix Transcription Dynamics Is Regulated by the X-to-Autosome Ratio and Semistable Transcriptional States. Mol Cell Biol 2016;36:2656-67. [Crossref] [PubMed]
- Chu C, Zhang QC, da Rocha ST, et al. Systematic discovery of Xist RNA binding proteins. Cell 2015;161:404-16. [Crossref] [PubMed]
- Chaligne R, Popova T, Mendoza-Parra MA, et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res 2015;25:488-503. [Crossref] [PubMed]
- Vincent-Salomon A, Ganem-Elbaz C, Manie E, et al. X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res 2007;67:5134-40. [Crossref] [PubMed]
- McHugh CA, Chen CK, Chow A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015;521:232-6. [Crossref] [PubMed]
- Yildirim E, Kirby JE, Brown DE, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 2013;152:727-42. [Crossref] [PubMed]
- Salvador MA, Wicinski J, Cabaud O, et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin Cancer Res 2013;19:6520-31. [Crossref] [PubMed]
- Fang J, Sun CC, Gong C. Long non-coding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun 2016;478:811-7. [Crossref] [PubMed]


