
IQGAP1 plays an important role in the tumorigenesis and invasion of papillary thyroid cancer
Introduction
Papillary thyroid cancer (PTC) is the most common type of thyroid malignancy, accounting for more than 80% of all cases of thyroid cancer. Moreover, the incidence of PTC has increased dramatically over the past several decades (1,2). Several genetic factors are involved in the development of PTC, including the activation of the MAPK signaling pathway as a result of a point mutation within BRAF, RAS mutation or RET/PTC rearrangement (3). Non-coding RNA (ncRNA), including small ncRNA and long ncRNA (lncRNA) has been revealed to be associated with the development of thyroid cancers. A number of studies have discussed the potential uses of ncRNA in the diagnosis and prognosis of thyroid tumors (4). Treatment for PTC with surgery in combination with radioiodine therapy is highly effective. Despite its relatively good prognosis with a 10-year cancer-specific survival rate above 90%, locoregional recurrences and distant metastasis still occur frequently (5). Accurate diagnosis of lymph metastasis and prediction of PTC recurrences is necessary to minimize poor outcomes.
IQ-domain GTPase-activating proteins (IQGAPs) are an evolutionarily conserved family of multi-domain proteins that are present in diverse organisms, ranging from yeast to mammals (6,7). The 189-kDa mammalian IQGAP1 protein, which has ubiquitous tissue expression, is the best-characterized member of the IQGAP family. IQGAP1 contains several protein-interacting domains, a calponin homology domain (CHD), a poly-proline-binding domain (WW), a calmodulin-binding motif (IQ motif), a Ras GTPase-activating protein (GAP)-related domain (GRD) and a Ras GTPase C-carboxy-terminal sequence (RGCT) (8). The functions of IQGAP1 protein include cytoskeletal regulation, coordinating cadherin mediated cell-cell adhesion, cell polarization and actin reorganization to promote cell migration (9). IQGAP1 binding partners are involved in several cell signaling pathways, including the mitogen- activated protein kinase (MAPK) cascade, the Wnt pathway, phosphoinositide 3-kinase (PI3K/Akt) signaling and other cell signaling processes affecting cell proliferation, differentiation and apoptosis (10,11). Accumulating evidence strongly supports a role for IQGAP1 in tumorigenesis and tumor invasion/metastasis. The established IQGAP1 binding partners coupled with the existing evidence on its role in neoplasms strongly suggest that IQGAP1 is an oncogene (12-14).
It has been proven that IQGAP1 protein plays a role in thyroid tumorigenesis (14,15). However, it is not well known whether IQGAP1 protein can be used as a diagnostic biomarker for PTC and/or whether it serves a role in promoting PTC cell proliferation, invasion and metastasis. Therefore, our objective in the current study was to identify the different IQGAP1 expression patterns in primary tumor, para-tumor and normal tissues and metastatic nodules. We also investigated its relationship with tumor proliferation, invasion and metastasis. Additionally, the potential regulatory mechanisms of IQGAP1 in PTC were explored by bioinformatics.
Methods
Patients and samples
Collected tumor samples included primary tumor, para-tumor and lymph nodule metastatic samples from PTC patients who underwent surgery at Xiangya Hospital of Central South University. These samples were divided into two groups, namely a with metastatic lymph nodules group (group 1: 16 tumor, para-tumor and lymph nodule metastatic tumor samples from PTC patients who underwent total thyroidectomy and regional lymph nodule dissection; lymph nodule metastases were verified during and after operation by pathological diagnosis) and a without metastatic lymph nodules group (group 2: 19 tumor and para-tumor samples from PTC patients who underwent total thyroidectomy, with no evidence of lymph nodule metastasis). A total of 13 normal tissues (normal thyroid tissues from patients who underwent benign nodules dissection) were used as a control group. This study was approved by the Ethics Committee of Xiangya Hospital, Central South University (Ethic Number: 201303192). Informed consents from patients were obtained permitting the use of their samples and publication of relevant data for the purposes of the present study.
Cell lines
The human PTC cell lines K1, BCPAP and TPC-1 and the normal cell line Nthy-ori 3-1 were used in this study. K1 cells were cultured in Dulbecco’s modified Eagle’s medium, and BCPAP, TPC-1 and Nthy-ori 3-1 cells were cultured in RPMI 1640, both supplemented with fetal bovine serum (10%).
Real-time quantitative PCR (qPCR)
The relative cDNA level of IQGAP1 in fresh tumor samples and cell lines was measured by qPCR using an ABI 7900 PCR Detection System with SYBR green dye (Applied Biosystems). The primer sequences for IQGAP1 were as follows: forward, 5'-AGAACGTGGCTTATGAGTACCT-3', and reverse, 5'-CCAGTCGCCTTGTATCTGGT-3'. Quantification of the relative IQGAP1 cDNA level, normalized to the internal control gene GAPDH, was performed using the 2-ΔΔCt method. The primer sequences for GAPDH were as follows: forward, 5'-TGACTTCAACAGCGACACCCA-3' and reverse, 5'-CACCCTGTTGCTGTAGCCAAA-3'. All samples were run in triplicate.
Western blotting
Briefly, the total proteins was separated on 15% SDS-PAGE gel, and then transferred to immunoblot PVDF membrane. The membranes were blocked with 5% defatted milk before being exposed to monoclonal anti-IQGAP1 antibody against human IQGAP1 (ABCAM, Cambridge, MA, USA; dilution 1:1,000). The blot was then incubated with a horseradish peroxidase-conjugated antibody. The experiment was performed at least 3 times.
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) blocks from tissue samples were cut into 5-µm thick tissue sections. The tissue sections were incubated with anti-IQGAP1 primary antibody (dilution 1:500) overnight at room temperature. Staining was visualized using DAB chromogenic substrate. Two independent pathologists blindly evaluated the immunostaining. Any differences in the interpretation of stained slides were resolved by consulting a third pathologist to achieve consensus. Totally, seven slides were reevaluated. The staining was considered positive if more than 10% of cells were stained strongly in the cytoplasm as described (16).
Cell culture and RNA interference (RNAi)
IQGAP1 expression in the cell lines was downregulated by transfection with small interfering RNAs (siRNAs) that were designed and synthesized by GenePharma Corp. (Shanghai, China). The commercial transfection reagent LipofectamineTM 2000 was used. The sequence of the four siRNAs used were IQGAP1-homo-1620 5'-GGCACAUGCAGAGAAUAAUTT-3' (siIQ1), IQGAP1-homo-3835 5'-GCCCACUUAAGCAUCAUUATT-3' (siIQ2), IQGAP1-homo-4665 5'-GCAGGUGGAUUACUAUAAATT-3' (siIQ3) and negative control: 5'-UUCUCCGAACGUGUCACGUTT-3' (siIQNC), respectively. A green fluorescent protein-tagged siIQNC was used as a control to determine transfection efficiency before the subsequent experiments. Western blotting and immunofluorescence staining assay were used to assess the success of transfection.
Immunofluorescence staining assay
Cells were placed into 6-well plain plates, and fixed in 4% formaldehyde and permeabilized in 0.5% Triton X-100. After incubation with primary antibodies at 4 °C overnight and appropriate secondary antibody tagged with a red fluorescent protein (Alexa Fluor 488), the cells were stained with 4',6-diamidino-2-phenylindole (DAPI) for 15 min. Specimens were examined using a LeicaTCS-SP5 fluorescence microscope.
MTT assays
Cell growth was determined by MTT assay (Sigma-Aldrich, USA). Cells were plated in 96-well plates at a density of 1×104 cells per well. At 12, 24, 48, 72 and 96 hours after siRNA transfection, 50 µL of 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to each well for 4 hours. The formazan crystals formed by viable cells were solubilized in DMSO and measured at 570 nm to determine absorbance values (expressed as the mean ± SD). The experiment was performed in triplicate.
Flow cytometry analysis
For the detection of cell proliferation, cells were fixed with 70% cold ethanol at 4 °C overnight at 48 hours after transfection. Then, 100 µL of RNaseA (1 mg/mL) and 100 µL of propidium iodide (100 mg/mL) were added into the cell suspensions and incubated at 37 °C or 30 minutes. Annexin V and 7-AAD stains were used to visualize apoptotic cells, according to the manufacturer’s instructions. The extent of cell proliferation and apoptosis was examined with an FC500 flow cytometer (Beckman Coulter, CA, USA). The cell proliferative index (PI) was determined as the sum of the cells in the S and G2/M phases of the cell cycle expressed as a fraction of the total cell population {PI = [(S + G2/M)/(G0/G1 + S + G2/M)] × 100%}.
Colony formation assay
Cells were trypsinized, centrifuged, resuspended, and plated onto agar at a density of 1×104 cells per well. After 14 days of incubation at 37 °C, the colonies were stained with 0.005% crystal violet at room temperature for 1 hour, and then assayed by visual counting in a blinded manner. The experiment was independently performed three times.
Scratch assay
Cells (transfected with siRNA or not) were grown to confluence in 96-well plates. A scratch was then made on the monolayer using a sterile pipette tip. Six fields per dish were photographed at 0, 8 and 24-hour points and the scratch area was measured. The experiment was repeated at least three times, and measurements were made in triplicate.
Transwell assay
A Transwell chamber (8-µm; Corning) was coated with Matrigel (Sigma) and inserted in a 24-well culture plate. Cells after transfection for 48 hours were resuspended and 5×104 cells were seeded in the upper chamber of each well. The migration assay was performed in 37 °C in a 5% CO2 tissue culture incubator overnight. The cells invading the Matrigel were fixed with 100% methanol and stained with hematoxylin. The number of cells was counted from three randomly selected visual fields using an inverted microscope at 200× magnification. The experiment was repeated three times.
Gene functional enrichment analysis by bioinformatics
Bioinformatics methods were utilized to identify the pathways that are significantly associated with upregulated expression of IQGAP1. The level 3 RNA sequencing data of PTC patients (including 507 PTC patients) in The Cancer Genome Atlas (TCGA) database was obtained from FireBrowse (http://firebrowse.org). According to the median expression values of IQGAP1, these patients were divided into an IQGAP1 high expression group and an IQGAP1 low expression group. The Limma package in R was used to identify differentially expressed genes (DEGs) between the two groups. We performed functional annotation clustering using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) online system. Gene Set Enrichment Analysis (GSEA) was also applied to determine enriched underlying biological pathways involved in the upregulated expression of IQGAP1.
Statistical analysis
The application of ggplot2 in R studio was used to draw the functional enrichment map. SPSS 21.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analysis of the experimental results. Student’s t-test and the Wilcoxon rank sum test were used to analyze the differences between data. P<0.05 was considered to indicate statistical significance.
Results
IQGAP1 expression is elevated in PTC tissues and cells
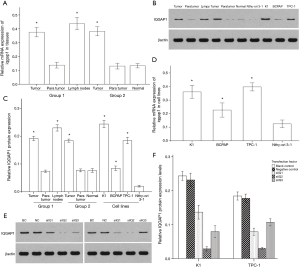
We detected the expression of IQGAP1 mRNA and protein in PTC tissues and cells by qPCR and western blotting. The levels of IQGAP1 mRNA and protein in cancer tissues were significantly higher than in para-tumor tissues and normal thyroid tissues (both in group 1 and 2). No significant differences in the mRNA and protein expression of IQGAP1 were observed between normal thyroid tissues and para-tumor tissues. Additionally, the highest expression level of IQGAP1 was observed in metastatic lymph nodules, while there was no difference in expression level between the primary tumors of each group (Figure 1A,B,C). The K1, BCPAP and TPC-1 cell lines exhibited higher IQGAP1 mRNA and protein expression levels than those of the normal cell line. Moreover, K1 and TPC-1 cells exhibited higher IQGAP1 expression levels than BCPAP cells (Figure 1D,E,F).
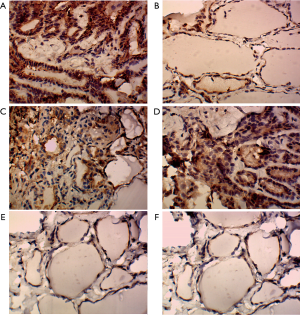
We further determined the expression of IQGAP1 in FFPE blocks from fresh tumor samples by immunohistochemical staining. Positive expression of IQGAP1, mainly located in the cytoplasm, was observed in 10 of 16 (62.5%) primary tumor tissues from group 1, 11 of 19 (57.9%) primary tumor tissues from group 2, and 12 of 16 (75%) metastatic lymph nodules, which exhibited intense positive staining (Figure 2). By contrast, positive expression of IQGAP1 was only observed in 2 of 13 (15.4%) normal tissues, 2 of 16 (12.5%) para-tumor tissues from group 1, and 2 of 19 (10.5%) para-tumor tissues from group 2 (Figure 2). These observations showed that IQGAP1 expression was upregulated significantly in PTC, especially in the metastatic lymph nodules.
RNA interference down-regulates IQGAP1 expression in PTC cells
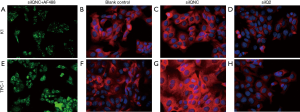
As the K1 and TPC-1 cells exhibited higher IQGAP1 expression than BCPAP cells, we performed our IQGAP1 knockdown studies using the K1 and TPC-1 cell lines. K1 and TPC-1 cells were transfected with siIQ1, siIQ2 and siIQ3, and siIQNC as a negative control. Non-transfected cells were used as blank control cells. At 24 hours after transfection, the transfection efficiency was over 80% (Figure 3). The results of a western blotting assay demonstrated that siIQ2 had the strongest inhibitory effects in both K1 and TPC-1 cells at a dose of 2 µmol/L (Figure 1E,F). Compared with the blank control cells, siIQNC cells showed no reduction in IQGAP1 expression levels, while siIQ2 cells exhibited a greater than 70% reduction in IQGAP1 expression levels (Figure 1E,F). Downregulation of IQGAP1 protein in K1 and TPC-1 cells was also visually examined by immunofluorescence staining (Figure 3). This confirmed that the transfection was highly efficient. Therefore, siIQ2-transfected K1 and TPC-1 cells were used in the subsequent experiments.
Gene silencing of IQGAP1 inhibits the proliferation of PTC cells
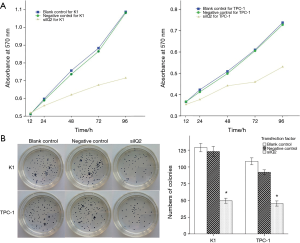
To confirm the functions of IQGAP1 in cell growth regulation, cell proliferation was monitored for 4 days after K1 and TPC-1 cells were transfected with siIQ2 or siIQNC as a control. During these 4 days, the growth rates of K1 and TPC-1 cells were reduced markedly. In addition, no differences were found between the blank and negative control cells over the entire experimental period (Figure 4A). A colony formation assay was used to assess the proliferative capacities of the cancer cells. The downregulation of IQGAP1 suppressed the colony-forming capacities of K1 and TPC-1 cells (Figure 4B). Specifically, the K1-siIQ2 and TPC-1-siIQ2 cells formed less colonies than the blank and negative control cells. Moreover, the cell numbers in each colony in the siIQ2 groups were significantly lower than those in the control groups (data not shown). A flow cytometry assay was subsequently performed to analyze cell cycle distribution profiles. The results showed that siIQ2 could significantly decrease the proliferation of K1 and TPC-1 cells, according to the percentages of S phase-cells and the PI values (Figure 5A). The downregulation of IQGAP1 expression caused cell cycle arrest, a decreased proliferation rate and an increased apoptosis rate in the K1 and TPC-1 cells (Figure 5B). These effects indicated that the silencing of IQGAP1 suppressed the growth and induced the apoptosis of PTC cells, suggesting that IQGAP1 is critical for thyroid carcinoma tumorigenesis and progression.
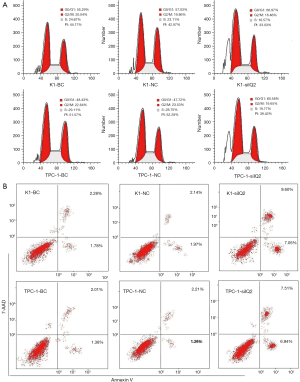
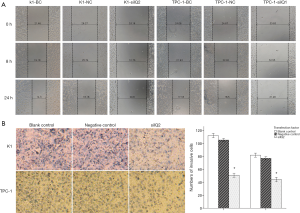
Down regulation of IQGAP1 protein inhabit PTC cellular invasion and migration in vitro
To examine whether the depletion of IQGAP1 had an effect on the migratory ability of the PTC cells, a wound-healing experiment was performed. As depicted in Figure 6A, cells infected with siIQ2 migrated more slowly and filled in the wounds at a slower rate than those in the siIQNC and blank control groups. The results were quantitatively assessed at 24 hours, and showed that the K1-siIQ2 and TPC-1-siIQ2 cells had notably low motility, indicative of retarded migration, compared with the blank and negative control cells (Figure 6A). In the invasion assay, we calculated the numbers of cells that migrated to the lower chamber side of the membrane. The data showed that there were an increased number of migrated K1 and TPC-1 cells in the blank control and siIQNC groups, compared with in the siIQ2 groups (Figure 6B). This demonstrated that IQGAP1 has the potential to enhance the invasive and metastatic abilities of PTC cells in vitro.
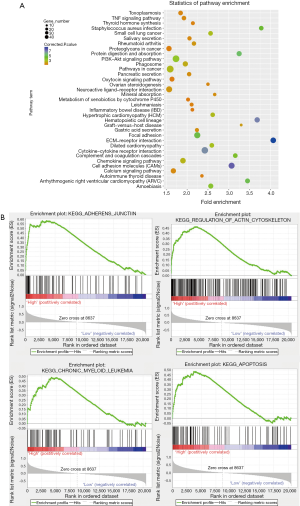
Identification of related pathways involved in the upregulation of IQGAP1
Differential analysis revealed 1,354 DEGs (865 upregulated and 489 downregulated) between the IQGAP1 high expression group and IQGAP1 low expression group. The DAVID online tool was used to classify these DEGs using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. The DEGs were significantly enriched in KEGG pathways including pathways in cancer, PI3K-Akt signaling pathway, cytokine-cytokine receptor interaction, focal adhesion, and cell adhesion molecules (ranked by the numbers of enriched genes, Figure 7A). GSEA is a technique that analyzes the overall expression trend of a group of genes to determine the differences between two biological states. We also carried out GSEA to identify tumor-related pathways associated with upregulated IQGAP1 expression. The topmost significantly enriched pathways related to upregulated IQGAP1 expression included adherens junction, regulation of actin cytoskeleton, chronic myeloid leukemia, apoptosis, JAK-STAT signaling pathways, and small cell lung cancer (Figure 7B). These results suggested that upregulated IQGAP1 expression was related to a series of important pathways and mechanisms involved in tumorigenesis and progression.
Discussion
IQGAP1 expression is frequently altered in different types of human tissues and neoplasms. Increased expression of IQGAP1 has been observed in several human neoplasms, including breast, colorectal, lung and liver cancer (12,17-19). In the present study, we demonstrated that IQGAP1 plays a critical role in PTC. Firstly, we found increased expression of IQGAP1 not only in primary PTC tissues, but also in PTC cell lines, and especially in metastatic lymph nodules. These results suggest that overexpression of IQGAP1 may contribute to the lymph node metastasis of PTC. For the first time, to the best of our knowledge, we also demonstrated the involvement of IQGAP1 in the proliferative and invasive capacities of PTC cells. Furthermore, we revealed that upregulated IQGAP1 expression was related to a series of important pathways and mechanisms involved in cancer through bioinformatics analysis. These findings suggest that IQGAP1 is involved in the tumorigenesis and progression of PTC, and are in accordance with the observations in other malignancies (9,12).
Recent studies have shown that IQGAP1 protein is involved in malignant tumors invasion/metastasis. IQGAP1 expression is related to the aggressive extent of malignancies (20-23), while a lack of IQGAP1 protein expression has been associated with a favorable prognosis in gastric cancer (24). The Rho family of GTPases largely coordinates the dynamic processes of cell migration and invasion. The scaffolding protein IQGAP1 binds to Cdc42, which has been characterized as a regulator of Rac1/Cdc42 signaling and actin dynamics (25,26). IQGAP1 cross-links actin filaments, calmodulin, and the APC microtubule-binding protein CLASP2, and thereby likely promotes cell migration (12,21,26). IQGAP1 is also involved in E-cadherin-mediated cell adhesion, and overexpression and a diffuse expression pattern of IQGAP1 at invasion fronts have been reported as independent prognostic parameters in carcinomas (13,17,27). In this study, there was no difference in the expression of IQGAP1 between primary tumors from the groups with or without lymph nodules metastasis. Interestingly, the metastatic lymph nodules exhibited the highest level of IQGAP1 expression. We believe that this unique expression pattern of IQGAP1 observed in our study suggests potential unknown mechanisms involving enhancement of IQGAP1 gene expression in malignant (metastatic) PTC cells. In other words, PTC cells with higher IQGAP1 expression levels may be more able to metastasize. In addition, we applied RNAi to specifically downregulate the expression of IQGAP1. Here for the first time, we demonstrated that reduced IQGAP1 expression attenuated the invasive/metastatic potential of PTC cells in vitro. Our data provide a possibility that IQGAP1 is an integral component of cell motility and participates in cell migration and invasion in PTC. Taken together, these data suggest that IQGAP1 overexpression is correlated with tumor progression. Nevertheless, the experiment in vivo will be needed to determine the feasibility of the current findings.
IQGAP1 participates in multiple essential aspects of mammalian biological processes; it mediates these effects by binding to and regulating the function of numerous interacting proteins. These interactions contribute to the functions of receptors and their cognate signaling cascade, with specific examples including influences on small GTPase function, cytoskeletal dynamics, neuronal regulation, intracellular trafficking, transcription, cell-cell adhesion, and cell migration (6,10,28). IQGAP1 is also present in a large cytoplasmic RNA-protein scaffold complex which contains lncRNAs, nuclear factor of activated T cells proteins (NFAT), and noncoding RNA repressor of NFAT (NRON), suggesting that IQGAP1 may participate in gene transcription via LncRNA-mediated regulation (29). The contribution of some of these cellular activities of IQGAP1 to different stages of cancer progression provides a clear link between IQGAP1 and cancer. In this study, two methods of pathway analysis were used to identify significant biological changes associated with the upregulation of IQGAP1 expression. We identified various pathways involved in cell proliferation, cell adhesion and apoptosis, and certain typical pathways associated with thyroid cancer including PI3K-Akt and JAK-STAT signaling pathways (3). These pathways were greatly associated with the upregulated expression of IQGAP1. Our study provides a comprehensive bioinformatics analysis of DEGs associated with the upregulation of IQGAP1, which may provide a set of useful pathways to investigate in the future, to further explore the molecular mechanisms underlying IQGAP1 involvement in PTC.
In summary, the present study demonstrated that upregulation of IQGAP1 in PTC tumors, especially in metastatic lymph nodules, may play critical roles in the tumor growth and lymph metastasis of PTC. These results suggest that IQGAP1 could serve as a biomarker for the diagnosis of malignant PTC. Furthermore, RNAi of IQGAP1 inhibited the growth and invasion of PTC cells, suggesting the potential therapeutic benefit of targeting IQGAP1 as a novel molecular marker of PTC progression. A better understanding of the mechanisms involved in these effects is now needed.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation (NSFC) of China (81372860, 81672885) and Innovative Foundation for graduate students of Central South University (2017zzts232).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.08.24). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Xiangya Hospital, Central South University (No. 201303192) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988-2005. Cancer 2009;115:3801-7. [Crossref] [PubMed]
- SEER Cancer Stat Facts: Thyroid Cancer. National Cancer Institute. Bethesda, MD. Available online: http://seer.cancer.gov/statfacts/html/thyro.html
- Buffet C, Groussin L. Molecular perspectives in differentiated thyroid cancer. Ann Endocrinol (Paris) 2015;76:1S8-15.
- Zhang R, Hardin H, Chen J, et al. Non-Coding RNAs in Thyroid Cancer. Endocr Pathol 2016;27:12-20. [Crossref] [PubMed]
- Wong KP, Lang BH. New molecular targeted therapy and redifferentiation therapy for radioiodine-refractory advanced papillary thyroid carcinoma: literature review. J Thyroid Res 2012;2012:818204 [Crossref] [PubMed]
- White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett 2009;583:1817-24. [Crossref] [PubMed]
- Shannon KB. IQGAP Family Members in Yeast, Dictyostelium, and Mammalian Cells. Int J Cell Biol 2012;2012:894817 [Crossref] [PubMed]
- Bashour AM, Fullerton AT, Hart MJ, et al. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J Cell Biol 1997;137:1555-66. [Crossref] [PubMed]
- Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cell Signal 2009;21:1471-8. [Crossref] [PubMed]
- White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal 2012;24:826-34. [Crossref] [PubMed]
- Brandt DT, Grosse R. Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep 2007;8:1019-23. [Crossref] [PubMed]
- Casteel DE, Turner S, Schwappacher R, et al. Rho isoform-specific interaction with IQGAP1 promotes breast cancer cell proliferation and migration. J Biol Chem 2012;287:38367-78. [Crossref] [PubMed]
- Guebel DV, Schmitz U, Wolkenhauer O, et al. Analysis of cell adhesion during early stages of colon cancer based on an extended multi-valued logic approach. Mol Biosyst 2012;8:1230-42. [Crossref] [PubMed]
- Liu Z, Liu D, Bojdani E, et al. IQGAP1 plays an important role in the invasiveness of thyroid cancer. Clin Cancer Res 2010;16:6009-18. [Crossref] [PubMed]
- Huang JK, Ma L, Song WH, et al. MALAT1 promotes the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. Biomed Pharmacother 2016;83:1-7. [Crossref] [PubMed]
- White CD, Khurana H, Gnatenko DV, et al. IQGAP1 and IQGAP2 are reciprocally altered in hepatocellular carcinoma. BMC Gastroenterol 2010;10:125. [Crossref] [PubMed]
- Hayashi H, Nabeshima K, Aoki M, et al. Overexpression of IQGAP1 in advanced colorectal cancer correlates with poor prognosis-critical role in tumor invasion. Int J Cancer 2010;126:2563-74. [PubMed]
- Zhao H, Xie C, Lin X, et al. Coexpression of IQ-domain GTPase-activating protein 1 (IQGAP1) and Dishevelled (Dvl) is correlated with poor prognosis in non-small cell lung cancer. PLoS One 2014;9:e113713 [Crossref] [PubMed]
- Xia FD, Wang ZL, Chen HX, et al. Differential expression of IQGAP1/2 in Hepatocellular carcinoma and its relationship with clinical outcomes. Asian Pac J Cancer Prev 2014;15:4951-6. [Crossref] [PubMed]
- Dong P, Nabeshima K, Nishimura N, et al. Overexpression and diffuse expression pattern of IQGAP1 at invasion fronts are independent prognostic parameters in ovarian carcinomas. Cancer Lett 2006;243:120-7. [Crossref] [PubMed]
- Zhou R, Guo Z, Watson C, et al. Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol Biol Cell 2003;14:1097-108. [Crossref] [PubMed]
- Huang L, Xu S, Hu D, et al. IQGAP1 Is Involved in Enhanced Aggressive Behavior of Epithelial Ovarian Cancer Stem Cell-Like Cells During Differentiation. Int J Gynecol Cancer 2015;25:559-65. [Crossref] [PubMed]
- Lu SH, Jiang XJ, Xiao GL, et al. miR-124a restoration inhibits glioma cell proliferation and invasion by suppressing IQGAP1 and beta-catenin. Oncol Rep 2014;32:2104-10. [Crossref] [PubMed]
- Walch A, Seidl S, Hermannstadter C, et al. Combined analysis of Rac1, IQGAP1, Tiam1 and E-cadherin expression in gastric cancer. Mod Pathol 2008;21:544-52. [Crossref] [PubMed]
- Watanabe T, Wang S, Noritake J, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell 2004;7:871-83. [Crossref] [PubMed]
- Watanabe T, Noritake J, Kakeno M, et al. Phosphorylation of CLASP2 by GSK-3beta regulates its interaction with IQGAP1, EB1 and microtubules. J Cell Sci 2009;122:2969-79. [Crossref] [PubMed]
- Schmidt VA. Watch the GAP: Emerging Roles for IQ Motif-Containing GTPase-Activating Proteins IQGAPs in Hepatocellular Carcinoma. Int J Hepatol 2012;2012:958673 [Crossref] [PubMed]
- Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep 2003;4:571-4. [Crossref] [PubMed]
- Sharma S, Findlay GM, Bandukwala HS, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A 2011;108:11381-6. [Crossref] [PubMed]


