
Gut flora shift caused by time-restricted feeding might protect the host from metabolic syndrome, inflammatory bowel disease and colorectal cancer
Introduction
Studies of the microbiome of the gut have gained increasing attention in the past decade. A healthy intestinal micro-environment can be characterized by considerable stability and diversity, idealized composition, and a desirable functional profile. The symbiotic interactions between the intestinal microbiota and the digestive tract both contribute to the maintenance of gut homeostasis. A wide spectrum of diseases, including obesity, metabolic syndrome, nonalcoholic steatohepatitis, inflammatory bowel diseases (IBD), atherosclerosis, type 1 diabetes, asthma (1), childhood undernutrition (2), autism, etc. (3,4), has been reported to have a close relationship with gut flora. A comprehensive understanding of diet-microbiota interplay could thus help identify a target for interventions aimed at reversing dysbiosis and restoring a healthy micro-ecosystem (1). The gut microbiota adapts itself to dietary regimen, which is itself determined by many factors, including geographic origin, host genotype, age, stress, antibiotics, and diet. These and a few other factors can change the general diversity of the microbiota and bloom specific bacterial groups, causing subsequent health outcomes (5). Gut flora shift also depends on the host’s individual response to dietary factors (6). Furthermore, enterotypes have been found to be closely related to long-term dietary habits, while changes induced by switching diet can be detected within 24 hours (7,8). Although the remarkable complexity of microbial diversity and function have proven to be a challenge to grasp fully, researchers have gradually gathered evidence revealing both the beneficial and deleterious effects of certain diets (9), along with their subsequent disease effects (10). Among this research, food-content studies have been the most common. For instance, a significant amount of research has established that proper dietary content, like fiber, is a key to curbing the pandemic of metabolic and digestive diseases via a boost to the colonic microbial synthesis of anti-inflammatory and anti-carcinogenic metabolites (9,11). Our own study, rather than focusing on content, concentrates on the feeding schedule as a critical aspect of diet, and does so through an investigation of the alteration of microbial diversity and functional alterations upon time-restricted feeding (TRF).
Methods
Animals
An animal experiment was conducted under the permission of the Animal Welfare Committee of Peking Union Medical College Hospital (permission no. XHDW-2015-1098), and performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines of the Salk Institute. Kunming male mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., 4 Yangshan Rd, Beijing, China) at 8 weeks of age were entrained to a 12:12 light-dark cycle. Intervention started after acclimatization with normal chow food available ad libitum (AD) for 1 week.
Diets and feeding schedules
Two feeding regimens were used in this study: mice under TRF were allowed access to food between zeitgeber time (ZT) 13 (ZT0: lights on, ZT 13: 1 h after lights off) and ZT21 (3 h before lights on), whereas mice under the AD regimen had access to food around the clock. In both regimens, caloric intake was unrestricted, and normal chow was used (GB14924.3-2010 Standard: 29% protein, 13% fat, 58% carbohydrate) (by reference: LabDiet-5010). Food was replenished at ZT13 each evening to assure an adequate amount of food was available for each cage. We randomly assigned 40, 9-week-old, wild-type Kunming mice into 4 groups: group A, which was treated with TRF for two months; group B which had AD access to food in the first month before TRF in the second; group C, which went through TRF in the first month but resumed AD pattern in the second month; and group D, which had continuous AD access to food.
Body weight
Body weight was monitored weekly.
DNA extraction and 16S rRNA sequencing
Ten mice (from 2 different cages) from each feeding group were sacrificed at ZT 21 after 2 weeks of feeding intervention. Ceca samples were isolated from the rectum and flash frozen. Each specimen was then resuspended and digested before lysis. DNA from the lysate was extracted, precipitated, washed, and resuspended. 16S rRNA gene sequence tags, corresponding to the hypervariable V1–V3 region, were generated using the 454 pyrosequencing platform. Alpha and beta diversity analyses were performed, and operational taxonomic unit (OTU)-based classification was used to generate data profiles.
Data analysis
Body weight comparison was performed by Stata/MP 14.0., and normally distributed data were displayed as mean ± s.e.m. Differences between the two groups were analyzed by the Student’s t-test. A value of P<0.05 indicated statistical significance.
Results
TRF attenuates weight gain (Figure 1)
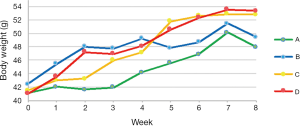
As all mice were randomly assigned to 4 groups, no difference in body weight was revealed after accommodation. At the end of week 4 (the first month after dietary intervention), the average weight of mice treated with the TRF regimen (group A and C) and those who had AD access to food (group B and D) were 45.67±0.64 vs. 48.67±0.63 g, respectively (n=20, P=0.002). From the beginning of week 5, group B was switched from the AD to the TRF feeding regimen, which explains the abrupt down-regulation of body weight. Likewise, the rapid increase in weight of group C can be attributed to the adjustment from the TRF to the AD mode in this group. At the end of week 8, average weights of group A, B, C, and D were 47.97±0.77, 51.66±1.34, 52.82±0.68, and 53.40±1.29 g, respectively (n=10). The duration of TRF implementation had a significant difference on body weight (group A versus B, P=0.0034), while a single month of TRF caused a decrease in body weight, but not significantly so (group B vs. D, P=0.14). Although the mean body weight of mice that began treatment with TRF and switched to AD (group C) was slightly lower than that of the group that followed AD for the whole duration of the study (group D), no statistical significance was shown (group C vs. D, P=0.34).
TRF causes notable changes in gut microbiota
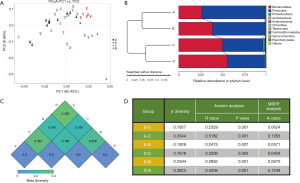
The figure of weighted PCoA (Figure S1A) is a visualization of the similarities or dissimilarities between each individual decided by the 2 most predominant factors. The coordinates in different colors represent mice subjects on different feeding regimens, and the distribution demonstrates that the mice within a given group shared relatively similar traits with the mice of a different group. Figure S1B is the weighted UniFrac distance describing the comparability of groups. It can be concluded that the groups undergoing an identical feeding regimen were relatively close to one another, regardless of the previous feeding pattern. At the time samples were collected, group A and B were under TRF regimen, while group C and D had AD access to food. The weighted heat map (Figure S1C) is further evidence showing the similarities between the groups, where higher values indicate lower similarities. It can be seen that the microbial ecosystem of group C and D shared many common features, whereas group A and D had the least homogeneity. Analysis of similarities (ANOSIM) and multi-response permutation procedure (MRPP) analysis are both statistical methods to evaluate the validity of grouping (Figure S1D). Regarding ANOSIM analysis, a positive R-value indicates that the deviation within groups is smaller than the one existing between groups. Our data set, with all figures being greater than 0 and all P value <0.01, shows that dietary timing is a strong intervention and not a random confounding factor. Likewise, the positive A value of MRPP analysis further attests to the notion that variations between groups were driven by different feeding schedules.
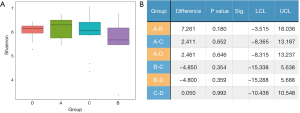
The Shannon index reflected α diversity of an ecosystem based on the weighted geometric mean of the proportional abundances of the types (Figure 2A). It was shown that no statistical significance was reached between any 2 groups. We also evaluated the abundance-based coverage estimators (ACE), Chao1, and Simpson index, yet the order of α diversity among the 4 groups were not accordant to each other using different analytical methods: (in descending order) Shannon: A-D-C-B; ACE: A-B-C-D; Chao1: A-B-C-D; Simpson: D-A-C-B. Meanwhile, all P values were greater than 0.05, which were considered not significant. Therefore, it must be concluded that no determinate evidence regarding the difference of α diversity among different groups was found (Figure 2B).
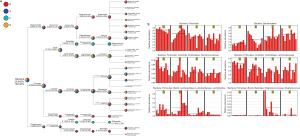
The taxonomy tree (Figure S2A) is an overview of the constitutions of gut flora of different groups, and clearly conveys the heterogeneity of bacterial composition through visualization. Although the highest hits under a given level might be similar, many significant variations merit attention as they have been proven to be closely related to the etiology and development of a wide variety of diseases. More detailed information is listed in Table 1. Linear discriminant analysis (LDA) combined with effect-size measurements (LEfSe) detects all possible features that could potentially explain the differences between different groups. Figure S2B captures some of the most noticeable results: the Firmicutes phylum was relatively abundant among feces of mice who were treated with TRF at the moment of specimen harvest (groups A and B); the Bacteroidetes phylum was more frequently cultured from AD groups (groups C and D); both Ruminococcaceae and Lachnospiraceae families accounted for a higher percentage among group A subjects; Lactobacillus species was also more abundant in group A, and Staphylococcus was predominantly discovered among mice from group C and D.

Full table
Discussion
TRF, by manipulating gene expression profiles, DNA repair, stem-cell renewal, and the restoration of blood-glucose homeostasis, attenuates fattening traits and metabolic disorders, improves cardiac function, and even lowers the risk of cancerous diseases (12). It is widely accepted that the feeding/fasting cycle influences host metabolism; however, little is known regarding the fundamental characteristics of the changes that happen in the gastrointestinal tract. Previous studies have claimed that diet-induced obesity dampens the feeding/fasting rhythm and diminishes many physical fluctuations. Interestingly, TRF restores this circadian clock, which protects against obesity and metabolic diseases. Relevant changes can be traced to the cyclical changes in the gut microbiome (12).
No specific diet has been shown to directly cause, prevent, or treat diseases (13), but our findings suggest that TRF is of great potential to the prevention of metabolic conditions and colorectal diseases. Through an array of analytic methods, we first confirmed that inter-group variation was definitively greater than intra-group variation. By the time of specimen harvest, groups on the same feeding pattern—group A and B, group C and D—were more phenotypically similar to each other than groups on a different feeding pattern. Group B and C did share some common traits as both groups both went through one month of TRF and one month of AD regimen. Microbiologists agree that the increase of α diversity or species richness in ecological terms indicates a healthier ecosystem (14). For example, in Crohn’s disease (CD) patients, reduced diversity can be discerned, even when inspecting the inflamed versus non-inflamed tissues within the same patient (15). Although no statistically significant difference in α diversity was found in our experiment, insights into the constitution of the gut flora in the 4 groups have very interesting implications.
In our study, bacteria groups proven to protect against the pandemic of obesity and metabolic syndrome were boosted by TRF. Some studies have conjectured that microbial biomarkers are potential signatures for personalized nutrition in the treatment of metabolic diseases (16). For instance, obese patients usually have more Actinobacteria and Bacteroidetes, but less Firmicutes, and activating the growth of Bacteroidetes and Actinobacteria simultaneously inhibits the growth of Firmicutes; this growth and concurrent inhibition may be responsible for the occurrence of obesity (17). Metagenomic analysis of the obese microflora shows that it is enriched for genes associated with lipid and carbohydrate metabolism (18). Our data illustrate that TRF increased the percentage of Firmicutes, while reducing those of Bacteroidetes and Actinobacteria (19). Another disease, diabetes mellitus, specifically type 2 diabetes, has positive relations with opportunistic pathogens, including Erysipelatoclostridium, which, in our study, was found to be less common in the TRF treated mice. Conversely, Butyrate-producing bacteria including Clostridiales, Lachnospiraceae, Ruminococcaceae, and Roseburia, which are all less frequent in diabetes cases (20,21), were shown to be the highest in group A, which was the group that underwent TRF for the longest period (2 months).
Dysregulation of the gut microbiome has also been implicated in the pathogenesis of colorectal diseases such as IBD and colorectal cancer (CRC). These two are the most comprehensively studied conditions that have been related most closely to the disruption of gut flora. Many studies have also reported lower biodiversity, known as α diversity, in IBD patients. Meanwhile, the other protective factors against IBD have also proven to be compromised in these patients, including the abundance of Bacteroides, Clostridia, Bifidobacteriaceae, and Ruminococcaceae (13,22-25). On the other hand, the increased numbers of adherent-invasive E. coli, Enterobacteriaceae, Fusobacteriaceae, etc., are associated with the disease state. Nowadays, microbiome-based diagnostics can distinguish pediatric patients with IBD from patients with similar symptoms (26). Although this test cannot replace endoscopy and histological examination as diagnostic tools, this non-invasive technique is an effective complement to the usual IBD detection suite. Mechanistic study shows that IBD pathogenesis may result from a dysregulation of the mucosal immune system, subsequently driving a pathogenic immune response against the commensal gut flora (27). Similarly, in our study, although α diversity failed to show statistical significance, the relative abundance of some taxa including the Clostridia class, Firmicutes phylum, Roseburia genus, and Ruminococcaceae family, provides evidence for TRF protection against IBD.
Other studies have yielded very similar results regarding the pathogenesis of colorectal cancer. Specifically, in vivo and in vitro data point to the beneficial effects of microbes in the families Lachnospiraceae, Ruminococcaceae, Prevotellaceae, and Lactobacillaceae, whereas families such as Enterobacteriaceae have been associated with an increased risk of colorectal cancer (7,28-31). Decreased relative abundance of butyrate-producing microbes from the families Ruminococcaceae and Lachnospiraceae and species such as Roseburia, Blautia and others, suggests a potential recurrence risk (32,33). In our study, the gut microbiota of TRF-regimen groups was highly consistent with the previous publications’ findings, giving us the confidence to announce the potential benefits of cancer prevention.
TRF protects the gut from disruption of the micro ecosystem through multiple biochemistry pathways. Disruption of the mucosal barrier by various endogenous and environmental agents exposes the host immune system to the resident microbiota, leading to proliferation of pathogen-specific and commensal-specific T cells (34). These cells migrate, react with the commensal microbiota and thus tip the balance from physiologic to pathologic inflammation. Butyrate, which is frequently mentioned in gut microbiome studies, has been shown to protect the bowel from colitis and CRC by lowering the oxidative damage to DNA, by triggering the occurrence of apoptosis of cells that are already damaged, by suppressing the growth of cancerous cells, and by decreasing the activity of co-carcinogenic enzyme (35). Moreover, in vitro studies have demonstrated that butyrate modulates the expression of the heat shock protein (HSP) 70, and caspase, which have important functions in apoptosis and inflammation (36,37). Other possible mechanisms modulated by butyrate are the modifying of pH and the adjustment of production for short-chain fatty acids (SCFAs) which are the primary energy source for colonic epithelial cells (especially acetate, propionate and butyrate) (38,39). SCFAs have recently been reported to induce the expansion of colonic Treg cells (40). Hence, generally speaking, butyrate-producing bacterial groups are important health-promoting factors. Overall, protective interventions, including TRF, decrease the propensity for oxidative stress, secretion of toxins, and virulence-related functions (41), which indicates a shift towards an inflammation-suppression microbiome (42).
If a thorough understanding of gut microbiota is achieved, the plasticity of the gut microecosystem can become a source of promising treatment options for diseases that have close relations to gut flora. While certain signatures drawn from gut microbiota have been used as powerful differential diagnostic methods (16,43), investigations highlighting diet, environment, gut microbiome, and physiological changes are also emerging (44,45). For instance, targeted bacterial microbiota modifications are innovative therapeutic strategies that may play a role in a wide range of diseases (46). Current studies have shown that certain traits are transmissible via fecal transplantation. Experiments specifically found that the colonization of germ-free mice with an “obese microbiota” resulted in a greater increase in total body fat compared to a colonization with a “lean microbiota” (47). However, the transplantation method crucially requires “healthy donors” in its implementation, which is a seriously rate-limiting step.
Accrued evidence provides potentially accurate information for manipulating the microbiota to promote health through diet or probiotics (48). Our study confirmed that not only does dietary composition affect the gut microbiota, but that eating schedule affects it as well. The fact that interactions play a role in health and disease, opens up the intriguing possibility of using dietary means to maintain an ideal health condition (16). Current studies have revealed the impact of a vegetarian diet on the microbiota and identified the subsequent fluctuation of certain species. Relationships between these changes and diseases like IBD have been indicated as well. With more and more pro-disease or pro-carcinogenic groups of microorganisms being specifically identified, it appears that the truth of disease onset and progression may hinge partially on the things that we eat (7).
Here we have proposed that TRF promotes anti-disease and anti-carcinogenic groups of microorganisms. Future investigations will open more therapeutic avenues of potential dietary interventions through manipulation of gut microbiota.
Acknowledgments
Funding: This study was supported by the CAMS Innovation Fund for Medical Sciences (2016-I2M-1-001) and the National High-tech R&D Program (863 Program) (2015AA020303).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.10.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted under the permission of the Animal Welfare Committee of Peking Union Medical College Hospital (permission no. XHDW-2015-1098), and performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines of the Salk Institute.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008;134:577-94. [Crossref] [PubMed]
- Hermann E, Foligne B. Healthy gut microbiota can resolve undernutrition. Hepatobiliary Surg Nutr 2017;6:141-3. [Crossref] [PubMed]
- Backhed F, Fraser CM, Ringel Y, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 2012;12:611-22. [Crossref] [PubMed]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol 2012;30:759-95. [Crossref] [PubMed]
- Konturek PC, Brzozowski T, Konturek SJ. Stress and the gut: pathophysiology, clinical consequences, diagnostic approach and treatment options. J Physiol Pharmacol 2011;62:591-9. [PubMed]
- Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 2011;5:220-30. [Crossref] [PubMed]
- Candela M, Turroni S, Biagi E, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol 2014;20:908-22. [Crossref] [PubMed]
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105-8. [Crossref] [PubMed]
- Vipperla K, O'Keefe SJ. Diet, microbiota, and dysbiosis: a 'recipe' for colorectal cancer. Food Funct 2016;7:1731-40. [Crossref] [PubMed]
- Teschke R, Schulze J. Green tea and the question of reduced liver cancer risk: the dawn of potential clinical relevance? Hepatobiliary Surg Nutr 2017;6:122-6. [Crossref] [PubMed]
- Pillai S. Rethinking mechanisms of autoimmune pathogenesis. J Autoimmun 2013;45:97-103. [Crossref] [PubMed]
- Zarrinpar A, Chaix A, Yooseph S, et al. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 2014;20:1006-17. [Crossref] [PubMed]
- Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014;146:1489-99. [Crossref] [PubMed]
- Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006;55:205-11. [Crossref] [PubMed]
- Sepehri S, Kotlowski R, Bernstein CN, et al. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis 2007;13:675-83. [Crossref] [PubMed]
- Korpela K, Flint HJ, Johnstone AM, et al. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One 2014;9:e90702 [Crossref] [PubMed]
- Barczynska R, Kapusniak J, Litwin M, et al. Dextrins from Maize Starch as Substances Activating the Growth of Bacteroidetes and Actinobacteria Simultaneously Inhibiting the Growth of Firmicutes, Responsible for the Occurrence of Obesity. Plant Foods Hum Nutr 2016;71:190-6. [Crossref] [PubMed]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480-4. [Crossref] [PubMed]
- Biedermann L, Brulisauer K, Zeitz J, et al. Smoking cessation alters intestinal microbiota: insights from quantitative investigations on human fecal samples using FISH. Inflamm Bowel Dis 2014;20:1496-501. [Crossref] [PubMed]
- Zhang D, Fu X, Dai X, et al. A new biological process for short-chain fatty acid generation from waste activated sludge improved by Clostridiales enhancement. Environ Sci Pollut Res Int 2016;23:23972-82. [Crossref] [PubMed]
- Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55-60. [Crossref] [PubMed]
- Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382-92. [Crossref] [PubMed]
- Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004;127:412-21. [Crossref] [PubMed]
- Ohkusa T, Sato N, Ogihara T, et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol 2002;17:849-53. [Crossref] [PubMed]
- Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292-8. [Crossref] [PubMed]
- Papa E, Docktor M, Smillie C, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 2012;7:e39242 [Crossref] [PubMed]
- Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest 2007;117:514-21. [Crossref] [PubMed]
- Zackular JP, Rogers MA. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112-21. [Crossref] [PubMed]
- Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett 2014;162:54-61. [Crossref] [PubMed]
- Arthur JC, Gharaibeh RZ, Muhlbauer M, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun 2014;5:4724. [Crossref] [PubMed]
- Ericsson AC, Akter S, Hanson MM, et al. Differential susceptibility to colorectal cancer due to naturally occurring gut microbiota. Oncotarget 2015;6:33689-704. [Crossref] [PubMed]
- Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 2013;66:462-70. [Crossref] [PubMed]
- Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013;8:e70803 [Crossref] [PubMed]
- Hand TW, Dos Santos LM, Bouladoux N, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 2012;337:1553-6. [Crossref] [PubMed]
- Rose DJ, DeMeo MT, Keshavarzian A, et al. Influence of dietary fiber on inflammatory bowel disease and colon cancer: importance of fermentation pattern. Nutr Rev 2007;65:51-62. [Crossref] [PubMed]
- Venkatraman A, Ramakrishna BS, Shaji RV, et al. Amelioration of dextran sulfate colitis by butyrate: role of heat shock protein 70 and NF-kappaB. Am J Physiol Gastrointest Liver Physiol 2003;285:G177-84. [Crossref] [PubMed]
- Brinkman BM, Hildebrand F, Kubica M, et al. Caspase deficiency alters the murine gut microbiome. Cell Death Dis 2011;2:e220 [Crossref] [PubMed]
- Yusuf F, Ilyas S, Damanik HA, et al. Microbiota Composition, HSP70 and Caspase-3 Expression as Marker for Colorectal Cancer Patients in Aceh, Indonesia. Acta Med Indones 2016;48:289-99. [PubMed]
- Ahmad MS, Krishnan S, Ramakrishna BS, et al. Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut 2000;46:493-9. [Crossref] [PubMed]
- Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013;500:232-6. [Crossref] [PubMed]
- Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [Crossref] [PubMed]
- Erickson AR, Cantarel BL, Lamendella R, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One 2012;7:e49138 [Crossref] [PubMed]
- Rossen NG, Fuentes S, Boonstra K, et al. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J Crohns Colitis 2015;9:342-8. [Crossref] [PubMed]
- Candela M, Biagi E, Maccaferri S, et al. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol 2012;20:385-91. [Crossref] [PubMed]
- Candela M, Biagi E, Turroni S, et al. Dynamic efficiency of the human intestinal microbiota. Crit Rev Microbiol 2015;41:165-71. [Crossref] [PubMed]
- Gagniere J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 2016;22:501-18. [Crossref] [PubMed]
- Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027-31. [Crossref] [PubMed]
- Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol 2015;50:495-507. [Crossref] [PubMed]


