
Potential roles of microRNA in levonorgestrel-treated uterine leiomyoma cells
Introduction
Uterine leiomyoma, a benign tumor, has an incidence rate of 30–50% (1) for women of a reproductive age, which increases to 70–80% when women are in their fifties (2). Generally, uterine leiomyoma is a major cause of hysterectomy and myomectomy. Uterine leiomyoma usually causes a cascade of gynecological damage, including menstrual abnormalities, recurrent pregnancy loss, pelvic pressure, etc. (3). Despite the high incidence of uterine leiomyoma, we still know little about its underlying etiology.
Previous studies have revealed that uterine leiomyoma originates from a myometrial cell transformation (4,5), while other studies have shown that the genes capable of encoding estrogen and progesterone receptors have been identified in the initiation and progression of leiomyoma (6-8). In some research, the increased risk of leiomyoma has been linked with genomic instability (9-11), implying microRNA (miRNA) potentially has a key role in leiomyoma (12).
MiRNAs, a type of small RNA with a length of around 20 nucleotides, might have a part in regulating gene expression. When binding with the 3'-untranslated region (3'-UTR) of target mRNAs in a sequence-specific way, miRNAs can induce target mRNA cleavage and degradation or inhibit translation by forming an RNA-interference-induced silencing complex (RISC) (6,7). miRNAs are also involved in multiple biological processes including cell growth, proliferation, differentiation and apoptosis. The majority of genes encoding miRNAs coincide with these cancer-related genomic regions, suggesting the potential role of miRNAs in tumorigenesis (7). There have been a few reports discussing miRNA dysregulation in uterine leiomyoma (8,9), but knowledge concerning the associations of miRNA functions and the therapy of uterine leiomyoma is still limited.
The levonorgestrel intrauterine system (LNG-IUS) is an intrauterine anti-fertility system, commonly used for uterine leiomyoma treatment. The LNG-IUS remedy has been found to reduce the size of uterine leiomyoma and relieve menstrual bleeding (2), yet the underlying mechanism still needs to be explored in depth. Here we aim to systematically analyze the miRNA expressions for the LNG-treated uterine leiomyoma cells in vitro. Hopefully, these results will facilitate a more comprehensive understanding of the biological mechanisms in the LNG-IUS remedy and offer new insights into effective clinical practice.
Methods
Isolation and primary culture of uterus leiomyoma cells
Inclusion criteria: patients with symptomatic leiomyomas, patients with regular cycle, patients who did not receive any hormone treatment for at least 3 months before hysterectomy, Uterus leiomyoma tissues were collected from 10 patients (mean age: 43.5±3.0 years old), who were scheduled to undergo hysterectomy. Surgery was performed 3 to 10 days after the cessation of menstruation. All of the leiomyomas selected for this study were 3–5 cm in diameter, and the tissues were all confirmed as uterine leiomyoma without degeneration. Leiomyoma cells were isolated using the digestion and tissue adherence method, and confirmed by the cell immunofluorescence with anti-α-smooth muscle actin antibody (5). The collected cells were embedded in Dulbecco’s Modified Eagle’s Medium (DMEM) (100 U/mL streptomycin, 100 U/mL penicillin, 10% heat-inactivated fetal bovine serum), cultured at 37 °C with 5% carbon dioxide under sustained humidity. The medium was replenished after digestion with trypsin, cultured for 24 hours (h) and then replaced every 48 h. Cell passage was performed when cell density went up to 95%. All the subsequent experiments were conducted within 6 generations from the primary leiomyoma cells. This study and related use of the patients’ samples were approved by the Ethics Committee of the Nanjing Maternity and Child Health Care Hospital Affiliated with Nanjing Medical University {IRB number: [2014] 72}, and all the participants gave informed consent before taking part.
Cell viability assay
A CCK-8 assay was performed to evaluate the effect of LNG on the UtLMC activity. To begin, 5,000 cells were seeded per well in a 96-well plate. Then, cells were treated with different doses of LNG (0, 5, 10, 15, 20, 25 ng/mL) (culture medium with equal amount of DMSO was taken as a solvent control) for 72 h. At last, CCK-8 detection agent was added, and the absorbance was read at 490 nm. The activity of the solvent control group was assumed to be 100%.
miRNA extraction for cultured cells
The total RNA of the cultured cells was isolated by miRNeasy Mini Kit (QIAGEN, Izasa, Barcelona, Spain), with the purity and concentration measured by NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE) and integrity examined by formaldehyde gel electrophoresis. The isolated total RNAs were fractionated and cleaned up using flashPAGE Fractionator and reagents (Invitrogen) (13) to extract miRNAs. Twenty-five µg of total RNA was loaded for flashPAGE purification. Targeted RNAs (~40 nucleotides) were tracked via a dye for isolation. Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA, USA) was employed to analyze the quality, yield and size of miRNA fractions and total RNA.
miRNA microarray analysis
A miRNA microarray analysis was performed via LC (Houston, TX, USA) in 3 independent groups: a solvent control group, an LNG-treated cell group (14), and a group of positive control (Ambion) composites of chemically synthesized oligoribonucleotides. miRNAs were labelled at 3’ amine-modified tails after isolation using the mirVana miRNA array labelling kit (Invitrogen) and coupled with Cy5 post-labelling reactive dye; then, a 3× miRNA hybridization buffer (Ambion) was added and heated at 95 °C for 2 min. The Cy5-labelled samples were cleaned using a post-labelling miRNA clean up kit, eluted, and stored at −70 °C or analyzed by hybridizing to miRNA arrays. On the micro-fluidic chip, each detection probe consisted of a chemically modified nucleotide sequence complementary to the target mRNA (from miRBase, http://microrna.sanger.ac.uk/sequences/v8.2) or other RNA control sequences, and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. After this, the mirVana miRNA Bioarray slides (Ambion) were placed into hybridization chambers (Corning Incorporated Life Sciences, Acton, MA) and 10 µL of miRNA were added to 20 µL of hybridization buffer mixture on the array slide under the Bioarray LifeSlip. The chambers were sealed and incubated in 42 °C water bath for 12~16 h. Each probe on the Bioarray slide was printed in duplicate with 20 positive and 100 negative controls.
miRNA target prediction and functional annotations
The potential targets of the differentially expressed miRNAs in the LNG-treated cells were predicted by the Target-Scan, PicTar, and microRNA.org programs with the default setting. Intersections of these miRNA target predictions were selected as miRNA targets. The MAS3 program (http://www.bioinfo.capitalbio.com/mas3/) was adopted to investigate the functions of the potential miRNA-targeted genes, in order to explore the biological functions for these candidate miRNAs. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were taken as annotation databases.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA (500 ng) was reverse-transcribed using the TaqManH MicroRNA Reverse Transcription kit (Applied Biosystems, Madrid, Spain), at 16 °C for 30 min, 42 °C for 30 min and 85 °C for 5 min, with the stem-loop primers provided by Ruibo (Guangzhou, China). The miRNA-specific cDNAs produced were amplified by PCR (TaqMan Universal PCR master mix from Applied Biosystems, Madrid, Spain; primers from Ruibo). qPCR was performed on the ABI 7900 PCR System with SYBR Green (Applied Biosystems, USA), at 95 °C for 10 min, 40 cycles for 15 s and 60 °C for 1 min, and replicated 3 times. The endogenous control was U6 small nuclear RNA. The relative expression was calculated by the 2−ΔΔCt method (15).
Statistical analysis
Microarray data were first subtracted from the background and then normalized by a LOWESS filter (Locally-weighted Regression) for the two-color experiments. Ratios for the 2 sets of detected signals (log2-transformed, balanced) and p-values of t-tests were calculated. The significance threshold was set at P<0.05, and the false discovery rate (FDR) was further calculated. The data was analyzed using the SPSS Graduate Pack 20.0 statistical software package (SPSS, Chicago, IL, USA). Intergroup differences were investigated by one-way analysis of variance. The significance of the GO term enrichment and the pathway correlated to the conditions, was determined by P values, in terms of EASE-score, Fisher P value or Hypergeometric P value.
Results
Inhibition of LNG on the viability of uterus leiomyoma cells
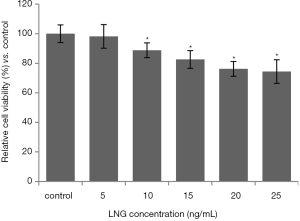
We performed a CCK-8 assay to evaluate the influence of LNG on uterus leiomyoma cell activities. The data showed that LNG (≥10 ng/mL) could significantly reduce cell viability, compared to the solvent control group. Also, the inhibition of LNG on cell viability appeared to be in a dose-dependent manner (Figure 1).
MiRNA expression signature in LNG-treated uterine leiomyoma cells
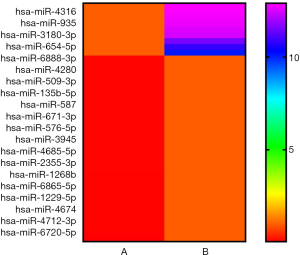
The microarray data illustrated that there were 722 up-regulated miRNAs and 740 down-regulated ones (absolute fold-change >2.0) in LNG-treated uterine leiomyoma cells. Compared to the solvent control group, 9 up-regulated and 32 down-regulated miRNAs had significantly changed expression after multiple testing correction [absolute fold-change >10.0, FDR <0.05] (Figure 2).
Discovery of miRNAs associated with LNG-treated uterine leiomyoma cells
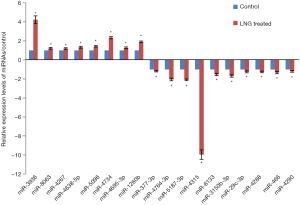
In this study, we identified 13 miRNAs that had significantly differential expression levels in LNG-treated uterine leiomyoma cells, compared with the solvent control. The qRT-PCR results showed that miR-1260b, miR-3936, miR-4267, miR-4638-5p, miR-4695-3p, miR-4734, miR-5096, and miR-8603 were significantly up-regulated in LNG-treated cells, while miR-377-3p, miR-29c-3p, miR-3150b-3p, miR-466, miR-4288, miR-4290, miR-4315, miR-4764-3p, miR-5187, and miR-6133 were significantly down-regulated (Figure 3).
GO and pathway analysis of differentially expressed miRNAs
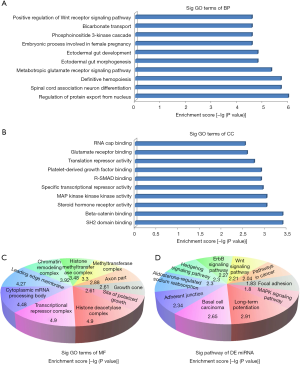
In order to explore what are the potential functions of the dys-regulated miRNAs in LNG-treated uterine leiomyoma cells, we predicted the targeted genes for those miRNAs through online bioinformatic tools. Next, we carried out GO analysis and found that these gene targets were mainly enriched in the following pathways: biological processes [i.e., positive regulation of Wnt receptor signaling pathway (GO: 0030177), bicarbonate transport (GO: 0015701), phosphoinositide 3-kinase cascade (GO: 0014065) (Figure 4A)]; molecular functions [i.e., RNA cap binding (GO: 0000339), glutamate receptor binding (GO: 0035254), translation repressor activity (GO: 0030371) (Figure 4B)]; and cellular components [i.e., histone deacetylase complex (GO: 0000118), transcriptional repressor complex (GO: 0017053), cytoplasmic mRNA processing body (GO: 0000932), leading edge membrane (GO: 0031256) (Figure 4C)]. Meanwhile, KEGG pathway analysis indicated several signaling pathways: long-term potentiation (hsa04720), basal cell carcinoma (hsa05217), adherens junction (hsa04520), aldosterone-regulated sodium reabsorption (hsa04960), hedgehog signaling pathway (hsa04340), ErbB signaling pathway (hsa04012), Wnt signaling pathway (hsa04310), pathways in cancer (hsa05200), focal adhesion (hsa04510), and MAPK signaling pathway (hsa04010) (Figure 4D).
Discussion
In the past decades, uterine leiomyoma has become the most common female reproductive system tumor. Usually, it is linked to ischemia, hypoxia, cellular injury, and myofibroblastic tumors, and can result in the unexpected expression of profibrotic, proinflammatory, and local angiogenic mediators (15-19). As advances in biological technology progress, efforts have been made to explore the mechanisms underlying the initiation and development of uterine leiomyoma. Non-coding RNAs have been proven to regulate protein coding gene expression and control some biological processes (20,21). MiRNA is one type of non-coding RNA, with its dys-regulation involved in various biological processes, including fibrosis, inflammation, and tumorigenesis (22,23). It has been confirmed that miRNAs are differentially expressed in the leiomyoma tissues compared with those expressed in normal tissues, but few miRNAs have been further investigated in relation to their function in regulating uterine leiomyoma (24-27). Therefore, our knowledge concerning the miRNAs involved in the leiomyoma therapy remains incomplete.
LNG-IUS has been established as an effective remedy to treat uterine leiomyoma. Thus, in this study, we selected LNG-treated cells as the experimental group, and solvent-treated cells as the control group. The results confirmed the effect of LNG on cell viability (Figure 1). Subsequently, we compared miRNA expressions via high-throughput miRNA microarray. We found 41 miRNAs were significantly dys-regulated in the LNG-treated uterine leiomyoma cells (Figure 2), indicating that these miRNAs participated in uterine leiomyoma therapy, and could be the potential targets for uterine leiomyoma treatment.
We validated the miRNA expression levels through qRT-PCR and replicated 13 miRNAs related to LNG treatment in uterine leiomyoma cells (Figure 3). Recent studies demonstrated that miRNAs could be the biomarkers or therapeutic targets for many diseases. In order to classify the potential functions of these 13 miRNAs, we carried out pathway analysis. Pathway analysis suggested that these gene products were mainly involved in the regulation of the Wnt receptor signaling pathway, bicarbonate transport, and phosphoinositide 3-kinase cascade (Figure 4A); these conclusions are also supported by other studies (28,29). Meanwhile, in line with Borahay et al.’s research (30), several KEGG signaling pathways were also involved, for example, long-term potentiation, basal cell carcinoma, adherens junction, aldosterone-regulated sodium reabsorption, hedgehog signaling pathway, ErbB signaling pathway, Wnt signaling pathway, pathways in cancer, focal adhesion, and the MAPK signaling pathway (Figure 4D).
There are still some limitations in the current study. First, the detailed molecular mechanisms have not been explored deeply. Second, the effects of other treatment modalities on the miRNAs expression have not been identified and compared with LNG treated leiomyoma cells. Without these comparisons, more significant insights into optimizing current therapy strategies cannot yet be proposed. Therefore, we will continue this work in future research.
Conclusions
In conclusion, we studied dys-regulated miRNAs in LNG-treated uterine leiomyoma cells. A network of differentially expressed miRNAs was constructed and numerous miRNAs were shown to participate in uterus leiomyoma therapy. However, in this iteration of the study, we did not examine the miRNA expression levels in INS-LNG treated patients. These dys-regulated miRNAs could prove to be the new therapeutic targets for uterus leiomyoma, and our observations here may warrant further studies on the pathogenesis of uterus leiomyoma.
Acknowledgments
We would like to thank the KangChen Bio-tech LTD (Shanghai, China), especially Feifei Xu (PhD) and Yajie Zhang for the miRNA microarray analysis.
Funding: This study was supported by the grants from the National Natural Science Foundation of China (No. 81202042, 81302304, 81402139, and 81401182), the Technology Development Foundation of Nanjing Medical University, China (No. 2017NJMUZD071, No. 2015NJMUZD063, No. 2015NJMUZD065).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.10.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study and related use of the patients’ samples were approved by the Ethics Committee of the Nanjing Maternity and Child Health Care Hospital Affiliated with Nanjing Medical University {IRB number: [2014] 72}, and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Laughlin-Tommaso SK, Jacoby VL, Myers ER. Disparities in Fibroid Incidence, Prognosis, and Management. Obstet Gynecol Clin North Am 2017;44:81-94. [Crossref] [PubMed]
- Baird DD, Dunson DB, Hill MC, et al. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol 2003;188:100-7. [Crossref] [PubMed]
- Kaganov H, Ades A. Uterine fibroids: Investigation and current management trends. Aust Fam Physician 2016;45:722-5. [PubMed]
- Rein MS, Barbieri RL, Friedman AJ. Progesterone: a critical role in the pathogenesis of uterine myomas. Am J Obstet Gynecol 1995;172:14-8. [Crossref] [PubMed]
- Stewart EA, Morton CC. The genetics of uterine leiomyomata: what clinicians need to know. Obstet Gynecol 2006;107:917-21. [Crossref] [PubMed]
- Vikhlyaeva EM, Khodzhaeva ZS, Fantschenko ND. Familial predisposition to uterine leiomyomas. Int J Gynaecol Obstet 1995;51:127-31. [Crossref] [PubMed]
- Luoto R, Kaprio J, Rutanen EM, et al. Heritability and risk factors of uterine fibroids--the Finnish Twin Cohort study. Maturitas 2000;37:15-26. [Crossref] [PubMed]
- Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: leiomyosarcoma. Cancer Genet Cytogenet 2005;161:1-19. [Crossref] [PubMed]
- Al-Hendy A, Salama SA. Catechol-O-methyltransferase polymorphism is associated with increased uterine leiomyoma risk in different ethnic groups. J Soc Gynecol Investig 2006;13:136-44. [Crossref] [PubMed]
- Denschlag D, Bentz EK, Hefler L, et al. Genotype distribution of estrogen receptor-alpha, catechol-O-methyltransferase, and cytochrome P450 17 gene polymorphisms in Caucasian women with uterine leiomyomas. Fertil Steril 2006;85:462-7. [Crossref] [PubMed]
- Villanova FE, Andrade PM, Otsuka AY, et al. Estrogen receptor alpha polymorphism and susceptibility to uterine leiomyoma. Steroids 2006;71:960-5. [Crossref] [PubMed]
- Chegini N, Verala J, Luo X, et al. Gene expression profile of leiomyoma and myometrium and the effect of gonadotropin releasing hormone analogue therapy. J Soc Gynecol Investig 2003;10:161-71. [Crossref] [PubMed]
- Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med 2015;19:2512. [Crossref] [PubMed]
- Ling J, Wu X, Fu Z, et al. Systematic analysis of gene expression pattern in has-miR-197 over-expressed human uterine leiomyoma cells. Biomed Pharmacother 2015;75:226-33. [Crossref] [PubMed]
- Wallez Y, Vilgrain I, Huber P. Angiogenesis: the VE-cadherin switch. Trends Cardiovasc Med 2006;16:55-9. [Crossref] [PubMed]
- Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol 2007;127:526-37. [Crossref] [PubMed]
- Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol 2008;214:199-210. [Crossref] [PubMed]
- Chegini N, Ma C, Tang XM, et al. Effects of GnRH analogues, ‘add-back’ steroid therapy, antiestrogen and antiprogestins on leiomyoma and myome-trial smooth muscle cell growth and transforming growth factor-beta expres-sion. Mol Hum Reprod 2002;8:1071-8. [Crossref] [PubMed]
- Fragiadaki M, Mason RM. Epithelial-mesenchymal transition in renal fibrosis - evidence for and against. Int J Exp Pathol 2011;92:143-50. [Crossref] [PubMed]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351-79. [Crossref] [PubMed]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011;12:99-110. [Crossref] [PubMed]
- Yang Y, Wang JK. The functional analysis of microRNAs involved in NF-kB signaling. Eur Rev Med Pharmacol Sci 2016;20:1764-74. [PubMed]
- Schickel R, Boyerinas B, Park SM, et al. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008;27:5959-74. [Crossref] [PubMed]
- Chuang TD, Khorram O. Mechanisms underlying aberrant expression of miR-29c in uterine leiomyoma. Fertil Steril 2016;105:236-45.e1. [Crossref] [PubMed]
- Qiang W, Liu Z, Serna VA, et al. Down-regulation of miR-29b is essential for pathogenesis of uterine leiomyoma. Endocrinology 2014;155:663-9. [Crossref] [PubMed]
- Chuang TD, Panda H, Luo X, et al. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr Relat Cancer 2012;19:541-56. [Crossref] [PubMed]
- Chuang TD, Khorram O. miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS One 2014;9:e95370 [Crossref] [PubMed]
- Ono M, Yin P, Navarro A, et al. Inhibition of canonical WNT signaling attenuates human leiomyoma cell growth. Fertil Steril 2014;101:1441-9. [Crossref] [PubMed]
- Karra L, Shushan A, Ben-Meir A, et al. Changes related to phosphatidylinositol 3-kinase/Akt signaling in leiomyomas: possible involvement of glycogen synthase kinase 3alpha and cyclin D2 in the pathophysiology. Fertil Steril 2010;93:2646-51. [Crossref] [PubMed]
- Borahay MA, Al-Hendy A, Kilic GS, et al. Signaling Pathways in Leiomyoma: Understanding Pathobiology and Implications for Therapy. Mol Med 2015;21:242-56. [Crossref] [PubMed]


