
Quantitative ubiquitylome analysis and crosstalk with proteome/acetylome analysis identified novel pathways and targets of perifosine treatment in neuroblastoma
Introduction
Neuroblastoma (NB) is an embryonic malignancy of the peripheral nervous system, derived from nerve-crest tissue and is mostly situated in adrenal medulla and paraspinal ganglia (1). As the most common pediatric extracranial solid tumor, NB accounts for almost 15% of cancer-related deaths during early childhood (2,3). According to its genetic heterogeneity, biological and clinical characteristics, NB patients are divided into low-, medium- and high-risk groups. Although clinical outcomes have improved during the last decade, the progress in high-risk NB patients is still limited with the long term survival rate only reaching 50%, despite therapeutic intensification with chemotherapy, radiotherapy, stem cell transplantation and immunotherapy (4,5). Thus, novel targets and effective therapeutic approaches are urgently needed for high-risk NB patients.
Perifosine is a third generation alkylphospholipid analog, which we have previously shown to suppress NB tumor growth both in vivo and in vitro (6). Moreover, our previous studies indicate that perifosine’s inhibition of AKT sensitizes NB cells to chemotherapy (7). Several clinical trials of perifosine have reported tolerable toxicity and prolonged progression-free survival in refractory high-risk NB patients (8,9). In a total of 27 patients who failed other treatments, 9 acquired long-term progression-free survival (from 43 to 74 months, median 54 months) with perifosine monotherapy. Interestingly, 8/9 responders were high-risk but had tumors which were non-MycN amplified (9). Frequently perifosine has been used as an oral AKT inhibitor, with little difference in activity in patients with or without PI3K mutation although it tended to be more effective in patients with a PTEN absence in a clinical trial of recurrent gynecologic cancer (10). These clinical trials suggest that in addition to AKT inhibition, perifosine is affecting other molecular mechanisms which may contribute to its therapeutic effect and AKT detection was not sufficient to specifically select patients for perifosine therapy. Thus, a more comprehensive understanding of the spectrum of mechanisms affected by perifosine is important to enhance its therapeutic effectiveness.
Proteomics is a convenient, high efficiency method to assess molecular changes in drug-related studies and assessments of post-translational modifications (PTM) offer insights into the regulation of protein function. Previously, we performed proteome and acetylome in a NB cell line-SK-N-AS (AS) cells after perifosine treatment (11). Ubiquitination, which refers to the process of conjugating ubiquitin to a lysine residue of a substrate protein, is another important type of PTM (12) and regulates the stability and activity of substrates. Mechanisms regulating ubiquitination have been implicated in NB tumorigenesis, differentiation, energy metabolism and apoptosis (13-16). To define ubiquitination alterations upon perifosine treatment, we first investigated the ubiquitylated proteome of NB cells through SILAC labeling and affinity enrichment followed by high-resolution LC-MS/MS analysis. Using bioinformatics analysis, comparison of the ubiquitylome with our previous proteome/acetylome data (11), revealed novel insights into the mechanism of action of perifosine in NB.
Methods
Cells and reagents
SK-N-AS (AS) cell line was cultured in RPMI-1640 medium (Pierce) with 10% fetal bovine serum (Gibco), 1% penicillin-streptomycin and 2 mM/L glutamine and sodium pyruvate at 37 °C in 5% CO2 incubator. We dissolved perifosine (Selleck) in dimethyl sulfoxide (DMSO) and stored it at 10 mM, −20 °C. Chemicals purchased from Sigma were DMSO, iodoacetamide (IAA), formic acid (FA), trifluoroacetic acid (TFA) and dithiothreitol (DTT). Deubiquitinase inhibitor PR-619 was purchased from Selleck.
SILAC labeling
Cells in control group and perifosine treatment group were labeled with heavy isotopic lysine and arginine (K6R10) and light isotopic lysine (K0R0) respectively, following instructions of SILAC Protein Quantitation Kit (Pierce, Thermo). After culturing for at least 6 generations, cells were treated with 10 µM perifosine or the same amount of DMSO for 16 h, washed twice with ice-cold PBS and then harvested.
Protein extraction and in-solution trypsin digestion
The harvested “heavy” and “light” labeled cells were sonicated 3 times on ice using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 M Urea, 5 mM DTT, 2 mM EDTA, 1.0% cocktail III and 50 µM PR619). The remaining debris was removed by centrifugation at 20,000 g at 4 °C for 10 min. After concentration measurement, equal amounts of crude proteins in the supernatant labeled “heavy” or “light” were mixed and the crude proteins were precipitated with TFA using a 15% final concentration (v/v) (soluble fraction). After washing twice with −20 °C acetone, the protein pellets were dissolved in 100 mM NH4HCO3 (pH 8.0) for trypsin digestion. Trypsin solution (Promega) (trypsin:protein =1:50) was added to proteins and then the protein pellets were digested at 37 °C for 16 hours. After alkylation reaction, trypsin (trypsin:protein =1:100) was added again and incubated (37 °C, 4 h).
HPLC fractionation and affinity enrichment
Tryptic hydrolysis was followed by separation of sample into 80 fractions through high pH reverse-phase HPLC using Agilent 300 Extend C18 column (5 µm particles, 4.6 mm ID, 250 mm length). The tryptic peptides were dissolved in NETN mixture (0.5% NP-40, 1 mM EDTA, 100 mM NaCl, 50 mM Tris-HCl, pH 8.0) and incubated with pre-washed antibody beads (PTM Biolabs, Hangzhou, China) overnight at 4 °C with gentle shaking. Peptides-enriched beads were washed 4 times with NETN and twice with ddH2O. The combined peptides were eluted from the beads with 0.1% TFA, collected, vacuum dried and cleaned using C18 ZipTips (Millipore).
LC-MS/MS analysis
Peptides were dissolved in 0.1% FA and directly loaded onto a reversed-phase pre-column (Acclaim PepMap 100, Thermo Scientific). To separate peptide fractions, a reversed-phase analytical column (Acclaim PepMap RSLC, Thermo Scientific) was used, with gradient of an increase in solvent B (0.1% FA in 98% ACN) from 6% to 22% for 26 min, 22% to 35% for 8 min, went up to 80% in 3 min and staying at 80% for another 3 min. The whole process happened at a constant flow rate of 300 nL/min on an EASY-nLC 1000 UPLC system. The following analysis of resulting peptides was carried out by Q ExactiveTM Plus hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific).
Database search
The data acquired from MS/MS scans were processed by MaxQuant with integrated Andromeda search engine (v.1.4.1.2). Tandem mass spectra were searched against SwissProt database (20,203 sequences) concatenated with reverse decoy database. Up to four missing cleavages, 4 modifications/peptides and 5 charges were allowed for trypsin/P as specified cleavage enzyme. We set mass errors of precursor ions and fragment ions to 10 ppm and 0.02 Da, respectively. Carbamidomethylation on Cys was taken as a kind of fixed modification while oxidation on Met, ubiquitination on Lys and N-terminal of a protein were considered variable modifications. As for protein, peptide and modification site, false discovery rate (FDR) thresholds were set at 1%. Minimum peptide length was set at 7. All the other parameters were set as default in MaxQuant. The probability of site localization was specified as >0.75.
Bioinformatics analysis
GO annotation was analyzed through the UniProt-GOA database and InterProScan soft. Wolfpsort was applied to predict subcellular localization. We utilized Kyoto Encyclopedia of Genes and Genomes (KEGG) database for protein pathways analysis, CORUM protein complex database for protein complex analysis, InterPro domain database for protein domain annotation and Motif-x for motif analysis. Heatmap was drawn by the “heatmap.2” of R-package. The search tool for the Retrieval of Interacting Genes/Proteins (STRING) database was used to outline protein-protein interactions. Two-tailed Fisher’s exact test was applied.
Co-immunoprecipitation and Western blotting
For detecting ubiquitination of endogenous HMGCR, AS cells were seeded at 1.5×106 per 100-mm dish on day 0. One day later, cells were treated with either 10µM perifosine or the same amount of DMSO for 16 h. AS cells were lysed with IP lysis buffer (SN002, Invent) supplemented with 1% cocktail and 50 µM PR-619. After protein extraction, protein concentration was determined using a BCA Protein Assay (P0011, Beyotime). Total proteins were first immunoprecipitated overnight at 4 °C with 3.35 µg HMGCR antibody (Abcam)/normal rabbit IgG (CST), followed by incubation of proteins with A/G agarose beads (Santa Cruz) overnight at 4 °C. Beads were washed with IP-lysis buffer 3 times and boiled at 100 °C for 5 min. Supernatants were loaded for SDS-PAGE gels and transferred to PVDF membranes (Millipore). Membranes were incubated with primary antibodies overnight at 4 °C. Quantification of Western blotting was performed by Chemical Luminescence Analyzer (Bio-Rad) with enhanced chemiluminescent reagents (Thermo Scientific). Primary antibodies included anti-HMGCR (Abcam, 1:5,000), and anti-ubiquitin (P4D1, 1:1,000). The antibody of ubiquitin (P4D1) was purchased from Cell Signaling Technology (CST) with product number as #3936. It is produced by immunizing animals with 1-76 full length ubiquitin. The antibody of ubiquitin can detect ubiquitin, polyubiquitin and ubiquitylated proteins.
Results
Overview of global ubiquitylome upon perifosine treatment in AS cell line
Our ubiquitylome analysis is based on the quantification of the di-glycine remnant (K-ɛ-GG), which is a distinct feature of ubiquitination (17). In this study, anti-K-ɛ-GG antibody (PTM Biolabs) was used to capture peptides containing K-ɛ-GG. Combined with SILAC, HPLC and LC-MS/MS, we quantified the ubiquitination of proteins in AS cells upon perifosine treatment. Overall, 3,974 lysine ubiquitination sites in 1,668 protein groups were identified, among which 3,935 sites in 1,658 proteins were quantified. By setting a differentially quantified threshold at >1.5, we acquired 297 up-regulated lysine ubiquitination sites in 216 proteins and 226 down-regulated lysine ubiquitination sites in 176 proteins. The top 10 positions with the greatest fold differences from either up-regulated or down-regulated group have been shown (Table 1). Since we found 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) in 2 of the top 10 positions in the up-regulated group, we chose HMGCR for further validation.
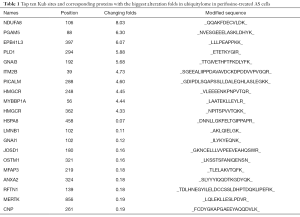
Full table
Motif analysis of ubiquitylome upon perifosine treatment in AS cells
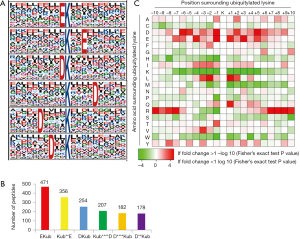
To specifically identify amino acids around ubiquitylated lysine sites, motif analysis was used and 6 motifs were prominently enriched: EKub, Kub**E, DKub, Kub****D, D****Kub and D**Kub (Kub represents the ubiquitylated lysine and * represents a random amino acid residue) (Figure 1A). The 6 motifs differed in abundance and EKub, Kub**E, DKub accounted for about 66% of all quantified peptides (Figure 1B). Heatmap based on the sequences of amino acids neighboring ubiquitination positions (Figure 1C) showed that aspartic acid (D) and glutamic acid (E) appeared most frequently around Kub sites while histidine (H) and methionine (M) seldom emerged.
Functional annotation and subcellular localization of differentially quantified Kub proteins
To investigate the features of the differentially expressed proteins after perifosine treatment, classification of gene ontology (GO) annotation and subcellular localization analysis were performed. Cellular process, single-organism process, biological regulation and metabolic process were leading categories when biological processes were evaluated (Figure 2A). In the cellular component analysis, cell and organelle related proteins stood out (Figure 2B). Molecular function analysis showed that the top 2 functions were binding (44%) and catalytic activity (28%) (Figure 2C).
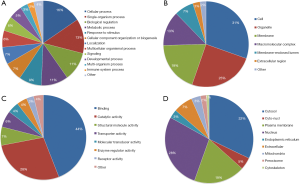
As for subcellular location prediction, ubiquitylation of proteins was distributed throughout the cell, with a slight majority in the cytoplasmic (32%), followed by the nucleus (28%) and plasma (19%) (Figure 2D).
Functional enrichment of differentially quantified Kub proteins
To explore the cellular functions of differentially regulated Kub proteins upon perifosine treatment, functional enrichment was conducted using GO, KEGG pathway, protein domain and protein complex.
In the ontology of cellular component, extracellular region, lipid particle and “extrinsic to membrane” were the most significantly enriched positions (Figure 3A the first 5 bars). For molecular function (Figure 3A the intermediate 5 bars), GTPase activity, phosphatidylinositol phosphate kinase activity and oxidoreductase activity were markedly enriched in perifosine-treated AS cells. Epidermal growth factor receptor (EGFR) signaling pathway, ERBB signaling pathway and salivary gland morphogenesis were the leading enriched processes (Figure 3A the last 5 bars).
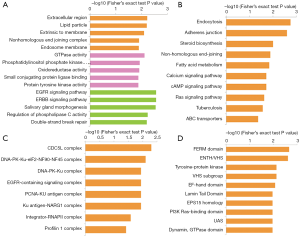
In the KEGG pathway analysis, endocytosis, adherens junction, steroid biosynthesis and non-homologous end-joining obtained remarkable enrichment after perifosine treatment (Figure 3B). In protein complex analysis, CDC5L complex, DNA-PK-Ku complex and EGFR-containing signaling complex were dominantly enriched (Figure 3C). Proteins containing FERM domain, VHS, tyrosine-protein kinase were prominently enriched in domain enrichment analysis (Figure 3D).
Functional enrichment-based cluster analysis of differentially quantified proteins
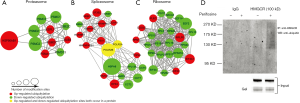
The protein-protein interaction network of ubiquitylome was established (Figure S1), followed by clustering ubiquitylated proteins into multiple biological processes (Figure 4A,B,C). It was observed that perifosine affected ubiquitination of proteins in the proteasome, spliceosome and ribosome, which indicates that perifosine might affect processes involved in gene transcription and translation.
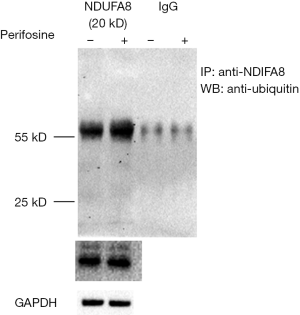
To validate the data of ubiquitylome, the antibody of HMGCR was used for immuno-precipitation and antibody of ubiquitin (P4D1) was used for western blotting to detect the alteration of HMGCR in ubiquitylated level (Figure 4D). HMGCR is the rate-limiting enzyme of cholesterol synthesis, which is of importance in cellular metabolism. The expression of ubiquitylated HMGCR was up-regulated after 10 µM perifosine treatment for 16 hours in AS cells, as evidenced by the high- molecular-weight smears of the immune-precipitates (Figure 4D). This was consistent with the results of quantitative ubiquitylome. Similar western analysis also validated ubiquitylation of NDUFA8 (Figure S2).
Crosstalk between global proteome and ubiquitylome
Based on the results of ubiquitylome in this study and the results of global proteome we reported previously (11), we found that 1,393 proteins in proteome also possessed quantified ubiquitination level in ubiquitylome, and 3,572 Kub sites in total were tested. We presented all data in a scatter plot to reflect the ratio of proteome and ubiquitylome. To eliminate the interference of protein expression, ubiquitination level was normalized and shown in red (Figure 5A). From the trend line of scatter plot (R2=−0.3938), proteome and ubiquitylome exhibited a slightly negative correlation after perifosine treatment. This result indicated that perifosine regulated protein expression partially through ubiquitination, such as ephrin type-A receptor 7 and protein sprouty homolog 1 (SPRY1). Yet there were some enzymes which showed a consistently changing trend in proteome and ubiquitylome, for example 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 3 (PFKFB3) and squalene synthase (FDFT1). This result indicated that besides protein expression, perifosine might also affect activity of enzymes through ubiquitination.
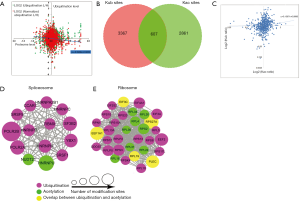
Several signaling pathways were enriched both in proteome and ubiquitylome analysis, including Ras signaling pathway, steroid biosynthesis, estrogen signaling pathway and endocytosis.
Crosstalk between quantitative ubiquitylome and acetylome
Ubiquitination and acetylation are both important PTMs. In present study, we compared ubiquitylome with previous acetylome (11) after perifosine treatment. Overall, 607 sites were modified both by ubiquitination and acetylation (Figure 5B). From the scatterplot, the two PTMs of those sites were not directly connected on the whole (R2=0.1387, Figure 5C).
We further performed protein-protein interaction network between ubiquitylome and acetylome (Figure S3) and totally 14 proteins were overlapped in differentially quantified proteins (Table 2). Subsequent cluster analysis showed that both ubiquitylome and acetylome were enriched in spliceosome (Figure 5D) and ribosome (Figure 5E). Certain proteins participating in the network were both ubiquitylated and acetylated, such as Eukaryotic translation initiation factor 3 subunit C (EIF3C), Elongation factor 1-alpha 1 (EEF1A1), Ubiquitin-40S ribosomal protein S27a (RPS27A), 60S ribosomal protein L (RPL), Plectin (PLEC). These proteins were likely to be important elements for perifosine treatment and therefore are preferential candidates for further study.

Full table
Discussion
Ubiquitination is a post-translational modification that is vital for the regulation of protein function and degradation. In the process of ubiquitination, target protein can be modified by mono-ubiquitin, multiple mono-ubiquitin, or polyubiquitin. There are 7 lysine residues (K6, K11, K27, K29, K33, K48 and K63) and an N-terminal methionine residue within a ubiquitin molecule that can all be targeted by ubiquitination to form different linkage types. The effect of ubiquitination varies with the linkage type of polyubiquitin chain and may affect protein cellular localization, protein-protein interactions and protein degradation by the 26S proteasome. As ubiquitination widely exists in living cells, it is involved in almost all cellular activities including cell signaling, endocytosis, DNA repair, and apoptosis (18-21). Therefore, we presumed that ubiquitination of the AS cellular proteome might also be affected by perifosine and probably be an underappreciated mechanism that contributes to its activity. Thus in this study, ubiquitylome was assessed and also compared with previous proteome/acetylome (11) in perifosine-treated AS cells.
In this study, we presented the top ten list of proteins with up/down regulated ubiquitination sites in AS cells upon perifosine treatment (Table 1). Among them, annexin A2 (ANXA2) is a member of the annexin family that exhibits Ca2+-dependent binding to phospholipids. ANXA2 was reported to play a role in the proliferation of astrocyte (22) which may affect permeability of the brain-blood barrier (23). Li et al. also found that ANXA2 was upregulated in glioblastoma tissues and associated with poor prognosis of glioma patients (24). EPB41L3 (Band 4.1-like protein 3), a marker of DNA methylation, has been implicated in the differentiation of embryonic stem cell into oligodendrocyte progenitor cells (25). In addition, EPB41L3 was relative with clinic-pathological characteristics of diffuse glioma patients (26). PGAM5 (Serine/threonine-protein phosphatase PGAM5) is a phosphatase that is involved in the respiratory chain of mitochondria. Researchers have found that PGAM5 is essential for head formation and the anterior-posterior body axis pattern via regulation of the Wnt/β-Catenin signaling pathway (27). Although functions of these proteins in NB are largely understudied, they all participate in the normal physiology of the nervous system and have pathologic consequences when disrupted.
In the motif analysis, we found the sequence of negative-charged amino acids [glutamic acid (E) and aspartic acid (D)] frequently emerged in the positions near Kub (Figure 1). This result was consistent with previous reports of ubiquitylome analysis in mammals (28,29) as well as in plants (30), indicating that it might be a general feature in lysine ubiquitination. As the spatial structure was altered in proteins undergoing ubiquitylation, the binding and corresponding catalytic activity would also be changed. This is consistent in our study as well as other reports in which the binding and catalytic activity compromised the largest share of the category of molecular function using GO annotation (28-30). In the functional clustering analysis of ubiquitylome after perifosine treatment, proteasome and ribosome were highly enriched (Figure 3A,B). A similar result was observed in the ubiquitylome study of tumor cells after treatment with the histone deacetylase suberoylanilide hydroxamic acid (SAHA) (31). Several proteins were both enriched upon SAHA and perifosine treatment such as 26S protease regulatory subunits (PSMC1, PSMC2, PSMC3, PSMD7 and PSMD13), heat shock 70 kDa protein (HSPA8) and 40S ribosomal protein S5. Meanwhile, the endocytosis and Ras signaling pathways were enriched in the KEGG pathway enrichment upon perifosine/SAHA treatment. The common points in the ubiquitylated proteome of perifosine and SAHA treated cancer cells suggest that the two drugs might share some similar mechanisms, indicating that perifosine might have a veiled mechanism other than as Akt inhibitor.
In our study, the Ras signaling pathway was prominently enriched both in global proteome and ubiquitylome after perifosine treatment. Since the Ras signaling pathway is upstream of PI3K-Akt in the signaling pathway (32,33), we presumed that perifosine might also affect the upstream targets of Akt in NB. The effect of perifosine on the Ras signaling pathway varies with cancer type (34,35) but is still unexplored in NB. Ras-related proteins circulate between active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate (GDP)-bound states. The factors stimulating the conversion from GDP-bound form to GTP-bound form are called guanine nucleotide exchange-factors (GEFs), and the reverse process is mediated by GTPase-activating proteins (GAPs) (36,37). In our study, the alterations in GEF (e.g., GTPase-activating protein 3) and GAP (e.g., Son of sevenless homolog) might be the key to the regulation of Ras signaling pathway by perifosine. Several effectors of Ras signaling pathway in our work have been studied in other cancers and might be potential targets in NB-including ADP-ribosylation factor 6 (ARF6) (38), Ras-related protein Ral-A (RALA) (39), Ras-related protein Rap-1A (RAP1A) (40), BRCA1-associated protein (BRAP) (41) and SPRY1 (42).
An enrichment of metabolic pathways in both global proteome and ubiquitylome indicated a potential impact of perifosine on metabolism. Carrasco et al. found disturbance of intracellular cholesterol transportation in perifosine-treated tumor cells (43). Perifosine inhibited cholesterol flux from plasma membrane to ER which led to cholesterol depletion in the ER, activation of transcription of cholesterogenic genes, causing accumulation of intracellular cholesterol (43). Therefore, we presumed that the activities of enzymes involved in cholesterol biosynthesis would be affected as well. HMGCR is the rate limiting enzyme of cholesterol synthesis. In our study, the level of ubiquitinated HMGCR was up-regulated perifosine treatment, as well as ubiquitin levels in several other cholesterogenic enzymes: Lanosterol 14-alpha demethylase (CYP51A1), SQLE, Methylsterol monooxygenase 1 (MSMO1), FDFT1 and Lanosterol synthase (LSS). SQLE was reported to predict poor prognosis and resistance to radiotherapy in prostate cancer (44) while FDFT1 was associated with poor prognosis and aggressive phenotype in prostate cancer (45). However, their function with enhanced ubiquitylation remained unknown and requires further study. Besides cholesterol metabolism, glycolysis is also very important in cancer cell metabolism (46). Thus the key enzymes and molecules participating in glycolysis have been intensively studied and previous studies revealed that active Akt was associated with enhanced glycolysis in cancer cells (47-49). As perifosine is an Akt inhibitor, there is evidence that perifosine impedes glycolysis and therefore improve chemo-sensitivity in non-small lung cancer cells (50). In a recently published study of human NB cells (SH-SY-5Y), researchers found that silencing of NADPH oxidase 4 inhibited glycolysis, and this was enhanced when combined with perifosine reversing hypoxia induced chemo-resistance (51). However, the underlying mechanism remains unclear. In the proteome and ubiquitylome of our study, PFKFB3, a key enzyme of glycolysis, is enriched after perifosine treatment. PFKFB3 was found up-regulated in several cancers (52,53) and levels decreased in cells exposed to Akt inhibitors (54), although perifosine was not evaluated.
Our extensive datasets detailing the ubiquitylome and crosstalk with proteome/acetylome reveals a number of targets and pathways previously unknown with perifosine treatment. Although the potential targets and pathways found in our present study need further confirmation, we believe that our study lays the basis to better understand the mechanisms of perifosine’s anti-tumor activities which will be important for a more precise application for patient’s therapy.
Acknowledgments
We thank the staff of the Medical Research Center of Shengjing Hospital and the PTM Biolabs for their support throughout these experiments.
Funding: This work was supported by the National Natural Science Foundation of China (No. 81472359, No. 81272538), the 2013 Liaoning Climbing Scholar Foundation, and the Natural Science Foundation of Liaoning Province (No. 201602855). Z Liu and CJ Thiele are supported by the Intramural Research Program of the NIH, Center for Cancer Research, NCI Grant Z01:ZIA BC 010789.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.11.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jiang M, Stanke J, Lahti JM. The connections between neural crest development and neuroblastoma. Curr Top Dev Biol 2011;94:77-127. [Crossref] [PubMed]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med 2010;362:2202-11. [Crossref] [PubMed]
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003;3:203-16. [Crossref] [PubMed]
- Maris JM, Hogarty MD, Bagatell R, et al. Neuroblastoma. Lancet 2007;369:2106-20. [Crossref] [PubMed]
- Park JR, Bagatell R, London WB, et al. Children's Oncology Group's 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer 2013;60:985-93. [Crossref] [PubMed]
- Li Z, Tan F, Liewehr DJ, et al. In vitro and in vivo inhibition of neuroblastoma tumor cell growth by AKT inhibitor perifosine. J Natl Cancer Inst 2010;102:758-70. [Crossref] [PubMed]
- Li Z, Oh DY, Nakamura K, et al. Perifosine-induced inhibition of Akt attenuates brain-derived neurotrophic factor/TrkB-induced chemoresistance in neuroblastoma in vivo. Cancer 2011;117:5412-22. [Crossref] [PubMed]
- Matsumoto K, Shichino H, Kawamoto H, et al. Phase I study of perifosine monotherapy in patients with recurrent or refractory neuroblastoma. Pediatr Blood Cancer 2017;64: [Crossref] [PubMed]
- Kushner BH, Cheung NV, Modak S, et al. A phase I/Ib trial targeting the Pi3k/Akt pathway using perifosine: Long-term progression-free survival of patients with resistant neuroblastoma. Int J Cancer 2017;140:480-4. [Crossref] [PubMed]
- Hasegawa K, Kagabu M, Mizuno M, et al. Phase II basket trial of perifosine monotherapy for recurrent gynecologic cancer with or without PIK3CA mutations. Invest New Drugs 2017;35:800-12. [Crossref] [PubMed]
- Gu X, Hua Z, Dong Y, et al. Proteome and Acetylome Analysis Identifies Novel Pathways and Targets Regulated by Perifosine in Neuroblastoma. Sci Rep 2017;7:42062. [Crossref] [PubMed]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998;67:425-79. [Crossref] [PubMed]
- Hagenbuchner J, Kiechl-Kohlendorfer U, Obexer P, et al. BIRC5/Survivin as a target for glycolysis inhibition in high-stage neuroblastoma. Oncogene 2016;35:2052-61. [Crossref] [PubMed]
- Emdal KB, Pedersen AK, Bekker-Jensen DB, et al. Temporal proteomics of NGF-TrkA signaling identifies an inhibitory role for the E3 ligase Cbl-b in neuroblastoma cell differentiation. Sci Signal 2015;8:ra40. [Crossref] [PubMed]
- Izumi H, Kaneko Y. Trim32 facilitates degradation of MYCN on spindle poles and induces asymmetric cell division in human neuroblastoma cells. Cancer Res 2014;74:5620-30. [Crossref] [PubMed]
- Cheng J, Fan YH, Xu X, et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis 2014;5:e1079 [Crossref] [PubMed]
- Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 2003;21:921-6. [Crossref] [PubMed]
- Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol 2008;15:20-2. [Crossref] [PubMed]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J 2005;24:3353-9. [Crossref] [PubMed]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007;315:201-5. [Crossref] [PubMed]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006;6:369-81. [Crossref] [PubMed]
- Chen J, Cui Z, Yang S, et al. The upregulation of annexin A2 after spinal cord injury in rats may have implication for astrocyte proliferation. Neuropeptides 2017;61:67-76. [Crossref] [PubMed]
- Li W, Chen Z, Yuan J, et al. Annexin A2 is a Robo4 ligand that modulates ARF6 activation-associated cerebral trans-endothelial permeability. J Cereb Blood Flow Metab 2018:271678X18777916. [Epub ahead of print].
- Li S, Zou H, Shao YY, et al. Pseudogenes of annexin A2, novel prognosis biomarkers for diffuse gliomas. Oncotarget 2017;8:106962-75. [PubMed]
- Chaerkady R, Letzen B, Renuse S, et al. Quantitative temporal proteomic analysis of human embryonic stem cell differentiation into oligodendrocyte progenitor cells. Proteomics 2011;11:4007-20. [Crossref] [PubMed]
- Perez-Janices N, Blanco-Luquin I, Tunon MT, et al. EPB41L3, TSP-1 and RASSF2 as new clinically relevant prognostic biomarkers in diffuse gliomas. Oncotarget 2015;6:368-80. [Crossref] [PubMed]
- Rauschenberger V, Bernkopf DB, Krenn S, et al. The phosphatase Pgam5 antagonizes Wnt/beta-Catenin signaling in embryonic anterior-posterior axis patterning. Development 2017;144:2234-47. [Crossref] [PubMed]
- Li W, Wang H, Yang Y, et al. Integrative Analysis of Proteome and Ubiquitylome Reveals Unique Features of Lysosomal and Endocytic Pathways in Gefitinib-Resistant Non-Small Cell Lung Cancer Cells. Proteomics 2018;18:e1700388 [Crossref] [PubMed]
- Kim W, Bennett EJ, Huttlin EL, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 2011;44:325-40. [Crossref] [PubMed]
- Guo J, Liu J, Wei Q, et al. Proteomes and Ubiquitylomes Analysis Reveals the Involvement of Ubiquitination in Protein Degradation in Petunias. Plant Physiol 2017;173:668-87. [Crossref] [PubMed]
- Wu Q, Cheng Z, Zhu J, et al. Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci Rep 2015;5:9520. [Crossref] [PubMed]
- Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer 2011;2:261-74. [Crossref] [PubMed]
- Castellano E, Sheridan C, Thin MZ, et al. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell 2013;24:617-30. [Crossref] [PubMed]
- Wu YL, Maachani UB, Schweitzer M, et al. Dual Inhibition of PI3K/AKT and MEK/ERK Pathways Induces Synergistic Antitumor Effects in Diffuse Intrinsic Pontine Glioma Cells. Transl Oncol 2017;10:221-8. [Crossref] [PubMed]
- Hideshima T, Catley L, Yasui H, et al. Perifosine, an oral bioactive novel alkylphospholipid, inhibits Akt and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Blood 2006;107:4053-62. [Crossref] [PubMed]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013;93:269-309. [Crossref] [PubMed]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007;129:865-77. [Crossref] [PubMed]
- Chabu C, Li DM, Xu T. EGFR/ARF6 regulation of Hh signalling stimulates oncogenic Ras tumour overgrowth. Nat Commun 2017;8:14688. [Crossref] [PubMed]
- Ginn KF, Fangman B, Terai K, et al. RalA is overactivated in medulloblastoma. J Neurooncol 2016;130:99-110. [Crossref] [PubMed]
- Bischoff A, Huck B, Keller B, et al. miR149 functions as a tumor suppressor by controlling breast epithelial cell migration and invasion. Cancer Res 2014;74:5256-65. [Crossref] [PubMed]
- Zhao Y, Wei L, Shao M, et al. BRCA1-Associated Protein Increases Invasiveness of Esophageal Squamous Cell Carcinoma. Gastroenterology 2017;153:1304-1319.e5. [Crossref] [PubMed]
- Koledova Z, Zhang X, Streuli C, et al. SPRY1 regulates mammary epithelial morphogenesis by modulating EGFR-dependent stromal paracrine signaling and ECM remodeling. Proc Natl Acad Sci U S A 2016;113:E5731-40. [Crossref] [PubMed]
- Carrasco MP, Jimenez-Lopez JM, Rios-Marco P, et al. Disruption of cellular cholesterol transport and homeostasis as a novel mechanism of action of membrane-targeted alkylphospholipid analogues. Br J Pharmacol 2010;160:355-66. [Crossref] [PubMed]
- Stopsack KH, Gerke TA, Andren O, et al. Cholesterol uptake and regulation in high-grade and lethal prostate cancers. Carcinogenesis 2017;38:806-11. [Crossref] [PubMed]
- Fukuma Y, Matsui H, Koike H, et al. Role of squalene synthase in prostate cancer risk and the biological aggressiveness of human prostate cancer. Prostate Cancer Prostatic Dis 2012;15:339-45. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Chae YC, Vaira V, Caino MC, et al. Mitochondrial Akt Regulation of Hypoxic Tumor Reprogramming. Cancer Cell 2016;30:257-72. [Crossref] [PubMed]
- Gong T, Cui L, Wang H, et al. Knockdown of KLF5 suppresses hypoxia-induced resistance to cisplatin in NSCLC cells by regulating HIF-1alpha-dependent glycolysis through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med 2018;16:164. [Crossref] [PubMed]
- Hagiwara A, Cornu M, Cybulski N, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 2012;15:725-38. [Crossref] [PubMed]
- Le Grand M, Berges R, Pasquier E, et al. Akt targeting as a strategy to boost chemotherapy efficacy in non-small cell lung cancer through metabolism suppression. Sci Rep 2017;7:45136. [Crossref] [PubMed]
- Yu T, Li L, Liu W, et al. Silencing of NADPH oxidase 4 attenuates hypoxia resistance in neuroblastoma cells SHSY-5Y by inhibiting PI3K/Akt-dependent glycolysis. Oncol Res 2018; [Epub ahead of print]. [Crossref] [PubMed]
- O'Neal J, Clem A, Reynolds L, et al. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) suppresses glucose metabolism and the growth of HER2+ breast cancer. Breast Cancer Res Treat 2016;160:29-40. [Crossref] [PubMed]
- Rodríguez-García A, Samso P, Fontova P, et al. TGF-beta1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J 2017;284:3437-54. [Crossref] [PubMed]
- Lu Q, Yan S, Sun H, et al. Akt inhibition attenuates rasfonin-induced autophagy and apoptosis through the glycolytic pathway in renal cancer cells. Cell Death Dis 2015;6:e2005 [Crossref] [PubMed]


