EML4-ALK translocation identification in RNA exosomal cargo (ExoALK) in NSCLC patients: a novel role for liquid biopsy
Introduction
Lung cancer treatment landscape has completely changed in the last decade. The introduction of druggable targets in the therapeutic scenario gave the opportunity to significantly improve the outcomes in the most lethal malignant disease. EML4-ALK translocation represents a story of success in drug development. The discovery of the aberration in tumors of patients with lung cancer in 2007 (1) soon translated into a rapid approval of the first ALK tyrosine kinase inhibitor (TKI), crizotinib (2), and the subsequent early arrival of second and third generation TKIs, all happening in roughly a decade. The current standard for the identification of the target necessarily implies identification of rearrangement in the tumor tissue either by FISH or immunohistochemistry. Unfortunately, despite the efforts, a big number of non-small cell lung cancer (NSCLC) patients have no tissue available for determination. The isolation of ctDNA is a novel approach, but still not validated for EML4-ALK translocation (3,4). Exosomes, new members of the liquid biopsy family, carrying genetic material represent an important tool for biomarkers discovery (5). Our group has identified, for the first time, EML4-ALK translocation in exosomes (ExoALK), using a next generation sequencing technique.
Material and methods
Patients selection
Patients with confirmed diagnosis of stage IIIB–IV NSCLC, naïve or under treatment with known ALK status were eligible for this proof of concept study. EML4-ALK translocation in tumor tissue was performed by FISH according to the clinical records of the patients. Patients gave their informed consent before the sample collection according to the institution regulation. Blood samples were collected only at a single point. Clinical data were collected from the electronic medical records. Samples and data from patients included in the study were provided by the Biobank of the University of Navarra (Reg#: B_0000612) and were processed following standard operating procedures approved by the Ethical and Scientific Committees. The trial was approved by the Antwerp University Hospital ethical committee n. 14/17/206.
Exosomal RNA isolation
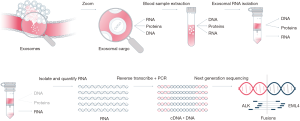
The exosomal RNA cargo was extracted from 1 mL of plasma using ExoRNeasy Kit from QIAGEN with a modification of the original protocol to avoid loss of material and volume. This includes 3 centrifugations of 5' × 500 g, 15' × 3,000 g and 30' × 10,000 g instead of the filtration in 0.8 μM (Figure 1).
Next generation sequencing
The fusions were analyzed using the Ion Ampliseq RNA Fusion Lung Cancer panel (Thermo Fisher Scientific, Waltham, USA). The primers used for amplification were then partially digested by the Pfu enzyme. The product of digestion was ligated with corresponding barcoded adapters and purified using Ampure Beads (Agilent Genomics Inc). The product of purification was amplified for 5 more cycles and subsequently purified using Ampure Beads. Ten pM of each library was loaded into the IonChef system (Thermo Fisher Scientific, Waltham, USA) for the emulsion polymerase chain reaction (PCR) and then loaded in the chip.
The quality of the data was assessed using the Torrent suite software (Thermo Fisher Scientific, Waltham, USA) associated with the sequencing machine. The minimum read length for fusion detection was set to 50 bp (base pair). We consider that under this value the reads were too short to overlap targeted fusion breakpoint. In addition, to assess the detection quality, at least 20,000 reads were required per sample (Figure 1).
Results
A total of 19 patients with plasma samples were included in the study, 16 of them harboring EML4-ALK translocations in tissue. In 1 mL of plasma, the RNA concentration ranged from 0.5 to 121 mg/mL. In total, 17 patients were finally eligible for extracellular vesicles analysis. In nine patients, we were able to identify the translocation in the exosomal RNA. The concordance between tissue and exosomes was 64% (9 out of 14 patients). All three patients being negative for the fusion gene in tissue resulted also negative in the ExoALK analysis, representing a specificity of 100% (Table 1). No correlation was found between the RNA yield and the type of sample or its clinical variables.
Table 1
| Exosomal RNA (ExoALK) (n=17) | Tissue (patients n=19) | |
|---|---|---|
| Positive | Negative | |
| Positive | 9 | 0 |
| Negative | 5 | 3 |
| Degraded RNA | 2 | – |
Sensitivity =64%; Specificity =100%.
Discussion
Here we demonstrate for the first time that EML4-ALK translocation detection in exosomes of NSCLC patients is feasible, with a high sensitivity and specificity. The exosomal analysis represents an opportunity for patients therapeutic selection with a minimal invasive procedure, easy to perform and with an affordable cost. The development of new techniques with a direct detection of the fusion by capture instead of by amplification would be the key to improve the sensitivity of ExoALK in the future.
Acknowledgments
The authors acknowledge OncoDNA for the free research collaboration, Dr. Maxime Lienard from OncoDNA for the contribution in exosomal cargo analysis and Prof. Patrick Pauwels from Molecular Pathology Unit at Antwerp University Hospital for the data analysis. Human samples and data from patients included in the study were provided by Biobank@UZA (Antwerp, Belgium; ID: BE71030031000), the AORN Vincenzo Monaldi, Department of Oncology, Naples, Italy and were processed following standard operating procedures approved by the Ethical and Scientific Committees. We particularly acknowledge the patients for their participation and the Biobank of the University of Navarra (Reg#: B_0000612) for its collaboration.
Funding: P Reclusa received a scholarship grant from University of Palermo.
Footnote
Provenance and peer review: The article was commissioned by the editorial office, Translational Cancer Research for the series “Targeted Therapy and Non-Small Cell Lung Cancer: A New Era?”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.11.35). The series “Targeted Therapy and Non-Small Cell Lung Cancer: A New Era?” was commissioned by the editorial office without any funding or sponsorship. UM and CR served as the unpaid Guest Editors of the series. The authors have no other conflicts of interest to declare.
Ethical statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). The trial was approved by the Antwerp University Hospital ethical committee n. 14/17/206. Informed consent was obtained from the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors Through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol 2018;2018: [Crossref] [PubMed]
- Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: The new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539-46. [PubMed]