
COPB2-shRNA suppresses cell growth and induces apoptosis of STAD SGC-7901 cells via inactivation of p38 MAPK/Smad2/Chk1/2 as an oncogene
Introduction
Stomach adenocarcinoma (STAD) is a common and lethal cancer over the world, with dramatic morbidity and mortality. A report of epidemiology data of gastrointestinal and liver cancers in the Asia-Pacific region showed that the incidence of STAD has declined significantly in the West but has increased in East Asia (1). Population-based cancer registration data of china in 2013 indicated that STAD incidences ranked the second in males’ population and the fourth in males population, mortalities ranked the third in males population and the second in males population (2). In spite of some developments in the treatment of STAD, the overall survival of STAD patients remains poor, since most STAD patients have local and distant metastases at diagnosis. Thus, it is of vital importance to find biomarkers and investigate relevant molecular mechanisms underlying vicious behaviors of STAD to provide novel targets for treatment. COPB2 is a subunit of the coatomer protein complex, a cytoplasmic protein complex that binds dilysine motifs and associates with Golgi non-clathrin-coated vesicles (3). The coatomer functions to mediate biosynthetic protein transport from the endoplasmic reticulum via the Golgi up to the trans-Golgi network. The coatomer is also required for Golgi membrane budding and is essential for the retrograde Golgi to endoplasmic reticulum transport of proteins. The coatomer is also involved in membrane trafficking in the exocytic pathway and for endocytic recycling of cell surface receptors. The association of the coatomer with the Golgi is regulated by binding ADP ribosylation factors and is GTP dependent. The coatomer sub-unit, which contains COPB2, can only be recruited when activated small guanine triphosphatases (GTPases) attract coat proteins to specific membrane export sites, thereby linking coatomers to export cargos. Recent researches indicated that COPB2 was a target gene of tumor target treatment with evidences in pancreas (4), colorectal cancer cell lines (4), prostate cancer cell line (5). Silencing COPB2 by lentivirus-mediated small interfering RNA (siRNA) would inhibit cell proliferation, arrest cell cycle at G1- and G2-phase (5), and even induce apoptosis. RNAi-mediated knockdown of COPB2 could inhibit growth and cellular invasion of lung cancer A549 cells significantly (6). However, there is still no report of COPB2 in STAD presently, it is extremely needed to investigate the function of COPB2 in STAD, such as cell proliferation, apoptosis and its mechanisms involved to uncover its role in STAD tumor genesis, and even find a new promising potential biomarker and target therapy gene.
Methods
Extraction and analyses of clinical data of COPB2
Expression data of COPB2 in STAD and clinical pathological characteristic data were collected from UALCAN (7) and GEPIA (8) web server (supported by TCGA database and the GTEx projects). COPB2 expression levels in STAD tissues and adjacent tissues, or other classification of pathological characteristic were compared and indicated as aberrant values of transcripts per million (TPM), and these pathological characteristic were as follows: expression of COPB2 in STAD based on tumor grade, H. pylori infection status, histological subtypes which could reflect clinical significances.
Cell lines and cell preparation
STAD cell lines, including AGS, MKN-45, MGC-803, BGC-823, SGC-7901, HGC-27, normal gastric epithelial cell line GES-1 and 293T cell line were provided by the Type Culture Collection of Cancer Institute and Hospital, Chinese Academy of Medical Sciences (CAMS) (Beijing, China). All cell lines were cultured in a medium containing DMEM and 10% heat-inactivated fetal bovine serum accompanied with 100 IU/mL Penicillin and 100 µg/mL Streptomycin. The cell culturing environment was prepared in an incubator having a humidified atmosphere of 5% CO2 at 37 °C. All cells reaching to exponential growing stage were used for further experiments.
Real-time quantitative PCR
Total RNA was separated from STAD cells and GES-1 cell line using RNAiso Plus (Takara Bio, Dalian, China). RNA purity and concentration were determined by P100+ UV-Vis spectrophotometer (Pultton, California, sunnyvale, United States). RNA was reversely transcribed using a Prime ScriptTM RT Reagent Kit (Takara Bio, Dalian, China). The reactions were performed by a Bio-Rad Real-Time PCR Detection System (ABI, ABI7500, USA) using a SYBR Master Mixture (Takara Bio, Dalian, China). The primers used were listed in Table 1. Each amplification reaction was executed in a tube with 20 µL volume containing 1 µL of cDNA, 0.5 mM of each primer and 1X SYBR Master Mixture. The amplification reaction program was composed of predenaturation step at 95 °C for 15 s, followed by denaturation and annealing steps of 40 cycles of 95 °C for 5 s, 60 °C for 30 s, in which fluorescence was collected. At the end of the amplification cycles, melting curve was generated to validate the specificity of the expected PCR product of COPB2. The expression levels of COPB2 mRNA were compared using normalization method with GAPDH, and the fold change was calculated using the 2−ΔΔCt method. Each sample was run in triplicates to get average value for analysis.

Full table
Detection of COPB2 mRNA expression in STAD cell lines
To understand the expression of COPB2 mRNA in COPB2 cell lines, Total RNA of STAD cell lines and GES-1 cells was extracted and reversely transcribed to cDNA, next, COPB2 was amplified in STAD cell lines and GES-1 cells, and the procedure were listed above. Following termination of PCR reaction, COPB2 mRNA expression was calculated and calibrated by GES-1 cells.
Lentiviral infection of SGC-7901 cells
Human STAD SGC-7901 cells were plated on six-well plates at 5×104 cells/well and incubated at 37 °C at 5×104 cells/well and incubated at 3 °C in 50 mL/L CO2 until 30% confluence was reached. The study was designed as both groups: negative control (shCtr, transfected with empty green fluorescent protein (GFP) lentivirus) and shCOPB2 group (shCOPB2, transfected with shCOPB2 GFP lentivirus). An appropriate amount of lentivirus was added according to the multiplicity of infection (MOI). The cells were repeatedly cultured in normal culture medium after 12 h. GFP-tagged gene expression was observed under a fluorescence microscope at 3 d after transfection, and cells with a transfection efficiency >80% were selected for subsequent analyses. Cells were harvested at 48 h after post-transfection for further analysis.
ShCOPB2 plasmid transfection in 293T cells
When the 293T cells were cultured and grew to the density of logarithmic growth phase, it was seeded in 24-well plate with 5×104/mL. Once reached to 80–90% confluence, the culture medium was replaced by opti-MEM1 of 400 µL for RNAi plasmid transfection. Next, the successfully constructed plasmids containing shCOPB2 or shCtrl of 0.5 µg and lipofectamine 2000 (Life Technologies, China headquarters of USA company, Shanghai, China) were dissolved in opti-MEM medium respectively and remained at room temperature for 5 minutes, and then, the plasmids and lipofectamine 2000 were mixed together and remained at room temperature for 20 minutes. After that, the mixture of plasmid DNA and Lipofectamine 2000 was added to 293T cells and cultured for 6–8 hours in incubator at 37 °C, 5% CO2 environment, and replaced with fresh complete culture medium containing 10% serum. The transfection rate was estimated by fluorescence microscope observation after 24 hours transfection.
Western blotting analysis of knockdown efficiency in 293T cells
After completed transfection for 36–48 hours, 293T cells of shCOPB2 and shCtr group were collected and lysed with the ice-cold lysis buffer for isolating total protein according to the manufacturer’s guide. After measured the protein concentrations using a Bio-Rad protein assay kit (Bio-Rad Laboratories, Shanghai, China), all protein samples were separated by 10% SDS-PAGE electrophoresis and transferred to PVDF membranes. Blots on PVDF membranes were incubated with the appropriate primary antibody (Mouse Anti-Flag, Sigma, 1:2,000, China headquarters, Shanghai, China) at room temperature. After washed in 5% non-fat milk in TBST (composed of Tris-Hcl, NaCl and tween20) saline at room temperature for 1 h, blots were incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (Goat Anti-Mouse IgG, 1:2,000, Santa-Cruz Biotechnology, Dallas, Texas, USA) for 1.5 h. Bands were evaluated using chemiluminescence (ECL, Thermo Scientific Pierce, Shanghai, China) and scanned images were quantified using ImageJ software (NIH Image for the Macintosh, USA). Western blotting experiments were performed in triplicate and the relative expression level of COPB2 was calculated using GAPDH (Mouse Anti-Flag, 1:2,000, Santa-Cruz Biotechnology, Dallas, Texas, USA) as a normalized control that is convenient to compare the relative expression level of COPB2 between shCOPB2 and shCtr group.
Cell proliferation
Following transfection, Cells of shCOPB2 and shCtr group were trypsinized and figured out the cell number. Cells (3,000 cells /well) were seeded in each well of 96-well plates in triplicate for each group and incubated at 37 °C for 24, 48, 72 and 96 hours. The cell counts were monitored over for these five 5 consecutive days. Prior to each termination time of cell culture in 5 consecutive days, 20 µL of 5 µmol/L MTT Formazan (Sigma-Aldrich, USA) was added and used to detect cell proliferation. The spectrophotometric absorbance of each sample was measured at 490 nm using a Benchmark microtiter plate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Cell counting by MTT and Cellomics Array Scan imaging detection
SGC-7901 cells of shCOPB2 and shCtr group were removed from plate using 0.25% trypsin-EDTA and suspended in standard medium when cells growth achieving logarithmic stage. Cells were seeded in five wells at 1000 cells/well, followed by further incubation at 37 °C and 50 mL/L CO2. A Cellomics Array Scan VT1 (Thermo Fisher Scientific) was used to monitor GFP expression of cells in both group continuously in each well over a 5-day period. In this study, statistical data were mapped and cell proliferation curves were drawn to compare the growth ability of SGC-7901 cells in both shCOPB2 and shCtr group.
Colony formation assay
Lentivirus-transduced shCOPB2 and shCtr cells were digested in 0.25% trypsin cells at 5×104 cells/mL. A hemocytometer was used to assess the cell count, and cell suspensions were transferred into six-well plates at 400–1,000 cells/well. The medium was replaced every three days for offering nutrition, and cell growth was observed usually. Till incubation lasted for 14 days or cells number in each colony in wells amounts to or more than 50, Colonies were visualized under a fluorescence microscope. Cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. The fully fixed SGC-7901 cells were stained with Giemsa (Dingguo Biotechnology Co., Ltd, Shanghai, China) 500 µL for 20 min, washed with ddH2O water and air-dried at room temperature. The total number of colonies with more than 50 cells was counted and images were captured using a light microscope and a fluorescence microscope. The assay was repeated three times.
Apoptosis assay
The cells were harvested with 0.25% trypsin from and washed once with 4 °C ice-cold D-Hanks (pH=7.2~7.4). Cells were centrifuged at 1,300 rmp for 5 min and rewashed with 1× binding buffer, and recentrifuged at 1,300 rmp for 3 min. The cells were then resuspended in 200 µL binding buffer to a final concentration of 106 cells/mL for subsequent analysis with the Annexin V-APC Apoptosis Detection Kit (eBioscience, San Diego, CA, United States) applied to assess apoptosis. The cell suspension of 100 µL volume was incubated with 10 µL Annexin V-APC in the dark for 15 min at room temperature. If needed, 400–800 µL 1× binding buffer was added to the stained cells according to the amounts of cells. The percentage of apoptotic rate was analyzed by flow cytometry. This assay was repeated in triplicate.
PathScan stress and apoptosis signaling antibody array assay
To detect the activation of possible intracellular signaling related to the influences in STAD induced by COPB2 silencing, the PathScan Stress and Apoptosis Signaling Antibody Array was used to screen and analysis relevant potential proteins. The procedure was briefed as follows, 5 days after lentivirus infection, SGC-7901 cells were collected and lysed. Intracellular signaling was detected using a PathScan® Stress and Apoptosis Signaling Antibody array kit (Cell Signaling Technology, Danvers, MA, USA) following the manufacturer’s instructions. This antibody Array is a slide-based antibody array which is upon the sandwich immunoassay principle used for detection of signaling nodes and downstream target nodes when phosphorylation activated at tyrosine or other residues. Detection of cells samples between COPB2 silenced group and that of ShCtr group was repeated in triplicate. Images were captured and analyzed to gather signals of aberrantly expressed protein targets by briefly exposing the slide to standard chemiluminescent film and were compared and calculated between two groups.
Statistical analysis
All experimental data were expressed as the means ± standard deviation from at least 3 separate experiments. Statistical analyses were evaluated using Student’s two-tailed t-test or analysis of variance by one-way ANOVA. Differences with P values of <0.05 are considered statistically significant.
Results
COPB2 was overexpressed in STAD tissue with clinical features
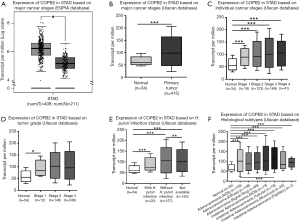
In order to discover the expression of COPB2 in STAD tissues and its potential clinical significances, we preliminarily analyzed the expression levels of COPB2 in STAD and adjacent tissues, in addition, other classification of pathological characteristic were also searched and analyzed by means of TCGA datasets through UALCAN and GEPIA web server. This result showed that COPB2 is upregulated in STAD tissues than in adjacent normal tissues (Figure 1A,B), and overexpression of COPB2 in cancer stage 1, 2, 3, 4 embodies more TPM values than normal tissues (Figure 1C) supported by both UALCAN and GEPIA web server. Overexpression of COPB2 in tumor stage1 embodies higher TPM value than in normal tissues (Figure 1D), overexpression of COPB2 in STAD tissues accompanied with or without H. pylori infection status, or not available status embodies more TPM values than in normal tissues (Figure 1E), overexpression of COPB2 in STAD tissues with histological subtypes also embodied higher TPM value than in normal tissues (Figure 1F). In summary, COPB2 is upregulated in STAD tissues, and which was associated with cancer stage 1, H. pylori infection and histological subtypes.
COPB2 was overexpressed in STAD cell lines
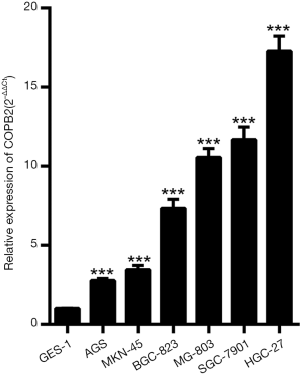
To investigate COPB2 expression in STAD cell lines, we detected its expression level by qRT-PCR in six STAD cell lines, including AGS, MKN-45, MGC-803, BGC-823, SGC-7901 and HGC-27, and the qRT-PCR results showed that the expression level of COPB2 mRNA in all six STAD cell lines is upregulated compared with GES-1 cell line (Figure 2).
ShRNA-mediated COPB2 knockdown efficiency in SGC-7901 and 293T cells
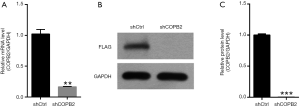
To investigate the effects of function loss of COPB2, we knocked down COPB2 in the SGC-7901 cells and 293T cells. STAD cell line SGC-7901 was cultured and successfully infected with Lv-shCOPB2 or Lv-shCtr with an infection rate greater than 80% for up to 72 hours after lentivirus infection. The mRNA expression level of COPB2 were significantly decreased in Lv-shCOPB2 group with a significant silenced efficiency amounts to 78.9% (P=0.0001) compared with Lv-shCtr group, as shown in Figure 3A. Human embryonic kidney 293T cells were transfected with shCOPB2 plasmid or shCtr plasmid, as shown in Figure 3B,C, Western blotting method detection showed that COPB2 protein expression was greatly reduced in the COPB2-shRNA infected cells compared with Lv-shCtr group (P=0.0023), indicating COPB2-shRNA could effectively silence COPB2 expression of the target sequence. These results demonstrated that the lentivirus-mediated targeting of COPB2 could effectively downregulate COPB2 expression in SGC-7901 cells and 293T cells at mRNA and protein levels respectively.
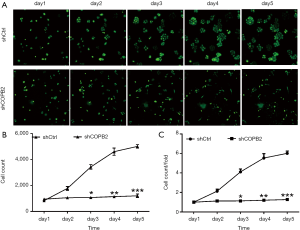
COPB2 silencing inhibited STAD cell proliferation by MTT and Cellomics Array Scan imaging detection
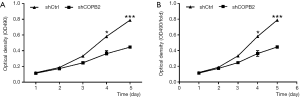
MTT results showed that the number and fold change of proliferation in SGC-7901 cells in Lv-shCOPB2 group were markedly reduced than that of cells in Lv-shCtrl group on the fourth and fifth day following COPB2 significantly silenced in SGC-7901 cells (P<0.05), as shown in Figure 4. Accordingly, the results suggested that knockdown of COPB2 will lead to the reduction of cell proliferation.
GFP-based cellomics array scan imaging assay showed that cell proliferation was significantly inhibited in COPB2-shRNA-silenced cells relative to cells of Lv-shCtrl group. The number of cells and the fold-change of proliferation were markedly reduced in the COPB2-shRNA-silenced STAD cells on the third, fourth and fifth day following COPB2 silenced significantly in SGC-7901 cells (P<0.05 or P<0.01 or P<0.001), as shown in Figure 5. Accordingly, the results suggested that downregulation of COPB2 could inhibit STAD cell proliferation.
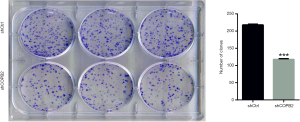
COPB2 silencing reduced STAD cells colony formation
Silencing of COPB2 reduced the anchorage-independent growth of STAD cell SGC-7901 in soft agar. The number of cell clones was significantly decreased in SGC-7901 cells infected with Lv-sh COPB2 (P=0.0005), as shown in Figure 6. The colony formation experiment confirmed that the silencing of COPB2 reduced the proliferative potential of SGC-7901 cells.
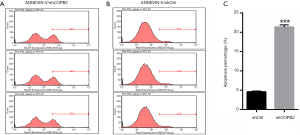
COPB2 silencing induced cell apoptosis
To elucidate whether downregulation of COPB2 could induce apoptosis in SGC-7901 cells, an apoptosis detection was designed to investigate the apoptotic rate between SGC-7901 cells of COPB2 silenced group and that of negative control cells. The apoptotic rate was assessed by flow cytometry using the Annexin V-APC Apoptosis Detection Kit. The proportion of apoptotic cells was significantly higher than that in SGC-7901-silenced cells than in the control cells, as shown in Figure 7. These data suggested that silencing of COPB2 could affect cell survival and induce apoptosis.
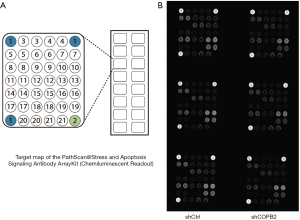
Apoptosis mechanism study of COPB2 silencing in STAD cells
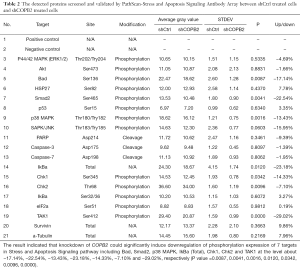
To investigate the modulation mechanism of COPB2 in the tumorigenesis of STAD, stress and apoptosis signaling pathway were analyzed in SGC-7901 cells after COPB2 knockdown. COPB2-triggered signal transduction was determined using the PathScan stress and apoptosis signaling antibody array kit. Knockdown of COPB2 significantly induced downregulation of phosphorylation expression of Bad, Smad2, p38 MAPK, IkBa (Total), Chk1, Chk2 and TAK1 at the level about −17.14%, −22.54%, −13.43%, −23.18%, −14.33%, −7.10% and −29.02%, respectively (P value=0.0087, 0.0041, 0.0016, 0.0120, 0.0342, 0.0096, 0.0000), as shown in Figure 8, Table 2. This result indicated that COPB2 could affect cell proliferation possibly via the phosphorylation—activated ATM/ATR-Chk1/Chk2 pathway and Ras/PI3K/Akt pathway. In addition, COPB2 silencing decreased the expression levels of Smad2, p38 MAPK, IkBa (Total), Chk1, Chk2 and TAK1 in STAD cells, indicating that Smad2, p38 MAPK, IkBa (Total), Chk1, Chk2 and TAK1 play important roles in the apoptotic process of SGC-7901 cells following COPB2 silencing. Furthermore, insightful studies are needed to clarify the mechanisms of COPB2 in STAD progression.
Full table
Discussion
In this study, we performed a clinical data analysis of COPB2 compared with normal tissues to discover its expression and clinical significances, and which was searched and extracted from UALCAN and GEPIA web server in STAD tissues. To be sure, COPB2 is upregulated in STAD tissues, and which was associated with cancer stage 1, H. pylori infection and histological subtypes. Therefore, it is suggested that overexpression of COPB2 in STAD tissues is likely to be a potential STAD biomarker and is worthy of be studied to explore its role, function and relevant mechanism in STAD cells. Subsequently, we found that the expression of COPB2 mRNA was significantly higher in all the tested human STAD cell lines compared to the normal gastric epithelium cell GES-1. In addition, silencing COPB2 inhibited proliferation, reduced cell colony formation and induced apoptosis of SGC-7901 cells. These data indicated that COPB2 overexpression play essential roles in the tumorigenicity of human STAD. These findings intensively suggested that COPB2 contributes to the pathogenesis of STAD through its proliferative and anti-apoptotic effects. Our data suggested that COPB2 overexpression may be an early event in gastric cell carcinogenesis. Therefore, COPB2 may represent a new promising target for gene therapy of STAD.
These findings suggested that shRNA mediated COPB2 knockdown result in a significant down-regulation of cell proliferation, colony formation, and a significant up-regulation of apoptosis in STAD cells. Our results were coincident with the outcome of COPB2 silencing in lung adenocarcinoma cells, in which cell growth was suppressed through apoptosis (6). From these data, it is suggested that COPB2 plays an essential role in STAD cell growth, indicating that the up-regulated expression of COPB2 has a positive impact on the progression of STAD cells and that shRNA-mediated gene silencing could effectively downregulate STAD progression in SGC-7901 cells.
Apoptosis has been considered as a principal role in anti-tumor strategy and designed to treat multiple cancers. There are multiple signaling pathways involved in the process of apoptosis; however, the pathways participate in apoptosis induced by COPB2 silencing still remained unknown. Therefore, this study was designed to analysis relevant signaling pathways using the Stress and Apoptosis Signaling Antibody Array comprehensively. The pathways and targets included in this array have been adopted to analysis relevant mechanisms of apoptosis, such as in glioblastoma cells (9), esophageal squamous cell carcinoma cells (10) and human bronchial epithelial cells (11). In this study, The results of Stress and Apoptosis Signaling Antibody Array assay showed that COPB2 silencing inhibited the activation of phosphorylation expression of Bad, Smad2, p38 MAPK, IkBa (Total), Chk1, Chk2 and TAK1. And these proteins represent essential molecular in multiple intracellular signal pathways.
Bad, an important molecular of the Bcl-2 family that normally binds to the Bcl-2/Bcl-extra larger (Bcl-xL) complex and could trigger cell apoptosis potentially, Once phosphorylation form of Bad dissociates from the Bcl-2/Bcl-xL complex, it could lead to the suppression of apoptosis. Bad of phosphorylation form at Ser-136, as a substrates of Akt, is always phosphorylated to promote cell proliferation and protein synthesis of human STAD cells, Downregulation p-Bad at Ser-136 is existed in apoptosis process (12). In addition, ERK could phosphorylate BAD at Ser-112 via p90RSK and promote cell proliferation of HeLa Cells and SH-SY5Y Cells (13,14), knockdown of ERK1 and/or ERK2 decreased p-Bad levels in the A375 melanoma cells (15). In this study, phosphorylation of p-Bad at Ser-136 is downregulated at about −17.14% in COPB2 silenced SGC-7901 cells compared with negative control SGC-7901 cells. This data indicated p-Bad at Ser-136 is downregulated after COPB2 silencing in human gastric adenocarcinoma SGC-7901 cells, and which proved apoptosis rates results detected by flow cytometry.
Chk1 and Chk2 kinases act as downstream targets of ATM and ATR signaling pathway, which belongs to the PI3/PI4-kinase family, respectively. Briefly, Chk1/Chk2 and upstream ATM/ATR are thought to be dominant controllers of cell cycle checkpoint signaling pathway, and which could respond to DNA damage and maintaining of genome stability, Chk2 is phosphorylated by ATM at residue Thr68 (16), and Chk1 is activated by the ATR-dependent phosphorylation of residue Ser345 (17). In this study, phosphorylation of Chk1 and Chk2 at Ser345 and Thr68 were downregulated at about 14.33% and 7.10% in COPB2 silenced SGC-7901 cells compared with negative control SGC-7901 cells respectively. This data suggested that the mechanisms of inhibition and apoptosis effects induced by silencing COPB2 is related to inactivation of ATM/ATR-Chk1/Chk2 signaling pathway.
Smad2, as a key mediator of TGF-β signaling, once stimulated by TGF-β, Smad2 phosphorylation at Ser465/467 would initiate and translocate into the nucleus (18). The outcome of TGF-β signaling is context dependent and can either induce apoptosis or contribute to tumor cell metastasis and epithelial-mesenchymal transitions in hepatoma cells (19,20). TAK1, as a kinase, it could be activated by TGF-β, bone morphogenetic proteins and other cytokines, Inhibition of both BMP and TGFβ signaling could promote the downregulation of TAK1 (21). TAK1 phosphorylates and activates the IKK complex composed of IKKα, IKKβ and NEMO (also called IKKγ), which leads to phosphorylation and degradation of IκB resulting in activation of NF-κB (22). Activated TAK1 also phosphorylates and activates MAPKKs leading to activation of MAPKs such as ERK, p38 and JNK (22). Phosphorylation of TAK1 at Ser412 responsible for activating NF-κB signaling and promotes aggressiveness of ovarian cancer cells, suppression of TAK1 activity would remarkably impair tumor growth and metastasis in ovarian cancer in vitro and in vivo (23). In this study, phosphorylation of Smad2 and TAK1 at Ser465 and Ser412 were downregulated at about −22.54% and −29.02% in COPB2 silenced SGC-7901 cells compared with negative control SGC-7901 cells. This data suggested that the mechanisms of growth inhibition and apoptosis effects induced by silencing COPB2 is related to inactivation of TGF-β-Smad2/TAK1 signaling pathway.
IκBα, a member of the NF-κB inhibitor family, could interact with Rel dimers to inhibit NF-κB/Rel complexes. Activation of NF-κB/Rel could occur through a proteasome-mediated degradation of IκBα and the inhibitor IκBα is targeted to its proteasome is mediated by phosphorylation of IκBα at Ser32 and Ser36 sites (24). In this study, phosphorylation of IκBα (Total) was downregulated at about −23.18% in COPB2 silenced SGC-7901 cells compared with negative control SGC-7901 cells. This data suggested that the mechanisms of growth inhibition and apoptosis effects induced by silencing COPB2 is related to downregulation of IκBα (Total) and activation NF-κB/Rel expression.
P38 MAPK is a member of the mitogen-activated protein kinase (MAPKs) family of serine/threonine protein kinases involved in many cellular processes such as cell proliferation, cell movement and cell death. The p38 MAPK signaling cascade is initiated when external stimuli such as growth factors, cellular stress. The mammalian p38 mitogen-activated protein kinases (MAPKs) family is composed of four members (p38α, p38β, p38γ, and p38δ), which are very similar in amino acid sequence but differ in their expression patterns, alternative p38MAPKs, p38γ and p38δ. These p38MAPKs show similarities to the classical p38α isoform and play an important role in certain physiological and pathological processes (25). Thr180 of p38alpha molecules is essential for catalysis, whereas Tyr182 is required for autoactivation and substrate recognition (26). In this study, phosphorylation of p38 MAPK at Thr180/Try182 is downregulated at about −13.43% in COPB2 silenced SGC-7901 cells compared with negative control SGC-7901 cells. This data indicated p-p38 MAPK at Thr180/Try182 is downregulated after COPB2 silencing in human gastric adenocarcinoma SGC-7901 cells. This data suggested that the mechanisms of growth inhibition and apoptosis effects induced by silencing COPB2 is associated with downregulation p-p38 MAPK at Thr180/Try182 and inactivation MAPK signaling pathway.
A previous study showed that COPB2, act as a potential synthetic lethal partners of KRAS, knocking down of COPB2 by siRNA induces a viability reduction of homozygous pancreatic KRAS mutated cell lines and heterozygous colorectal cancer cell line, downregulate the expression of p-Akt, p-Erk, p-Stat3, Xiap, Bcl-xl, and upregulate the expression of LC3I and LC3, which suggested that COPB2 influences KRAS signaling, apoptosis and autophagy by inactivating downstream RalGDS signaling including the small GTPases. RALA and RALB are activated which play a major role in vesicular trafficking and tumor proliferation (4). Protein microarrays or antibody array is an easily applicable tool to study disease by sandwich immunoassays with the features of high-throughput, high sensitivity and stringency (27-29), and upon these advantages, antibody array usually be employed in diagnosing inflammatory diseases, screening tumor biomarkers, and quantitatively detecting phosphorylation protein targets (30-32). In this study, a Stress and Apoptosis Signaling Antibody Array was utilized to screen and validate aberrant protein expression of phosphorylation form with sandwich immunoassay method linked by enzyme and amplified signals through biotin binding streptavidin tagged by HRP. As an effective method, this antibody array found 7 targets including Bad, Smad2, p38 MAPK, IkBa (Total), Chk1, Chk2 and TAK1 participate in growth inhibition and apoptosis, which responsible for COPB2 silencing effects in SGC-7901 cells.
Collectively, knockdown COPB2 could inhibit STAD cell proliferation and induce apoptosis, and the mechanisms of which may occur, in part, via the blockade of phosphorylation expression of Bad, Smad2, p38 MAPK, IkBa(Total), Chk1, Chk2 and TAK1.
Acknowledgments
Funding: This program was support by the 60th Project of China Postdoctoral Foundation (No. 2016M602888) and the Foundation of the fundamental scientific research funds for colleges and universities in Gansu Province [No. (2014)63-15].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.12.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Goh LY, Leow AH, Goh KL. Observations on the epidemiology of gastrointestinal and liver cancers in the Asia-Pacific region. J Dig Dis 2014;15:463-8. [Crossref] [PubMed]
- Zheng R, Zeng H, Zhang S, et al. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer 2017;36:66. [Crossref] [PubMed]
- Stenbeck G, Harter C, Brecht A, et al. beta'-COP, a novel subunit of coatomer. EMBO J 1993;12:2841-5. [Crossref] [PubMed]
- Christodoulou EG, Yang H, Lademann F, et al. Detection of COPB2 as a KRAS synthetic lethal partner through integration of functional genomics screens. Oncotarget 2017;8:34283-97. [Crossref] [PubMed]
- Mi Y, Yu M, Zhang L, et al. COPB2 Is Upregulated in Prostate Cancer and Regulates PC-3 Cell Proliferation, Cell Cycle, and Apoptosis. Arch Med Res 2016;47:411-8. [Crossref] [PubMed]
- Erdogan E, Klee EW, Thompson EA, et al. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res 2009;15:1527-33. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102 [Crossref] [PubMed]
- Li J, Ren S, Liu Y, et al. Knockdown of NUPR1 inhibits the proliferation of glioblastoma cells via ERK1/2, p38 MAPK and caspase-3. J Neurooncol 2017;132:15-26. [Crossref] [PubMed]
- Li B, Li YM, He WT, et al. Knockdown of DDX46 inhibits proliferation and induces apoptosis in esophageal squamous cell carcinoma cells. Oncol Rep 2016;36:223-30. [Crossref] [PubMed]
- Xu YM, Wu DD, Zheng W, et al. Proteome profiling of cadmium-induced apoptosis by antibody array analyses in human bronchial epithelial cells. Oncotarget 2016;7:6146-58. [PubMed]
- Zhang HY, Yang W, Lu JB. Knockdown of GluA2 induces apoptosis in non-small-cell lung cancer A549 cells through the p53 signaling pathway. Oncol Lett 2017;14:1005-10. [Crossref] [PubMed]
- Hafeez S, Urooj M, Saleem S, et al. BAD, a Proapoptotic Protein, Escapes ERK/RSK Phosphorylation in Deguelin and siRNA-Treated HeLa Cells. PLoS One 2016;11:e0145780 [Crossref] [PubMed]
- Meng XB, Sun GB, Wang M, et al. P90RSK and Nrf2 Activation via MEK1/2-ERK1/2 Pathways Mediated by Notoginsenoside R2 to Prevent 6-Hydroxydopamine-Induced Apoptotic Death in SH-SY5Y Cells. Evid Based Complement Alternat Med 2013;2013:971712 [Crossref] [PubMed]
- Qin J, Xin H, Nickoloff BJ. Specifically targeting ERK1 or ERK2 kills melanoma cells. J Transl Med 2012;10:15. [Crossref] [PubMed]
- Matsuoka S, Rotman G, Ogawa A, et al. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A 2000;97:10389-94. [Crossref] [PubMed]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 2001;21:4129-39. [Crossref] [PubMed]
- Souchelnytskyi S, Tamaki K, Engstrom U, et al. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem 1997;272:28107-15. [Crossref] [PubMed]
- Yoshimoto T, Fujita T, Kajiya M, et al. Involvement of smad2 and Erk/Akt cascade in TGF-beta1-induced apoptosis in human gingival epithelial cells. Cytokine 2015;75:165-73. [Crossref] [PubMed]
- Zhao HW, Li YW, Feng R, et al. TGF-beta/Smad2/3 signal pathway involves in U251 cell proliferation and apoptosis. Gene 2015;562:76-82. [Crossref] [PubMed]
- Augeri DJ, Langenfeld E, Castle M, et al. Inhibition of BMP and of TGFbeta receptors downregulates expression of XIAP and TAK1 leading to lung cancer cell death. Mol Cancer 2016;15:27. [Crossref] [PubMed]
- Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ 2014;21:1667-76. [Crossref] [PubMed]
- Cai PC, Shi L, Liu VW, et al. Elevated TAK1 augments tumor growth and metastatic capacities of ovarian cancer cells through activation of NF-kappaB signaling. Oncotarget 2014;5:7549-62. [Crossref] [PubMed]
- Yazdi S, Naumann M, Stein M. Double phosphorylation-induced structural changes in the signal-receiving domain of IkappaBalpha in complex with NF-kappaB. Proteins 2017;85:17-29. [Crossref] [PubMed]
- Risco A, Cuenda A. New Insights into the p38gamma and p38delta MAPK Pathways. J Signal Transduct 2012;2012:520289 [Crossref] [PubMed]
- Askari N, Beenstock J, Livnah O, et al. p38alpha is active in vitro and in vivo when monophosphorylated at threonine 180. Biochemistry 2009;48:2497-504. [Crossref] [PubMed]
- Gonzalez RM, Seurynck-Servoss SL, Crowley SA, et al. Development and validation of sandwich ELISA microarrays with minimal assay interference. J Proteome Res 2008;7:2406-14. [Crossref] [PubMed]
- Yu X, Schneiderhan-Marra N, Hsu HY, et al. Protein microarrays: effective tools for the study of inflammatory diseases. Methods Mol Biol 2009;577:199-214. [Crossref] [PubMed]
- Pla-Roca M, Leulmi RF, Tourekhanova S, et al. Antibody colocalization microarray: a scalable technology for multiplex protein analysis in complex samples. Mol Cell Proteomics 2012;11:M111.011460.
- Zhang Y, Liu W, Ge S, et al. Multiplexed sandwich immunoassays using flow-injection electrochemiluminescence with designed substrate spatial-resolved technique for detection of tumor markers. Biosens Bioelectron 2013;41:684-90. [Crossref] [PubMed]
- Gogalic S, Sauer U, Doppler S, et al. Investigating Colorimetric Protein Array Assay Schemes for Detection of Recurrence of Bladder Cancer. Biosensors (Basel) 2018;8: [Crossref] [PubMed]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. Sandwich assay for phosphorylation of protein multiplexes by using antibodies and Phos-tag. Anal Biochem 2013;438:104-6. [Crossref] [PubMed]


