
Expression levels and the prognostic value of long non-coding RNA PVT1 in serum of Han and Uygur gastric cancer patients in Xinjiang, China
Introduction
Gastric cancer (GC) is the second most common cause of cancer death worldwide and GC mortality in China makes up 42% of the general mortality rate of the world (1). Xinjiang is the largest autonomous region in northwestern China. The population in this region is made up of a number of ethnic groups. The major ethnic groups are the Han (39.7%) and the Uygur (45.7%). Different ethnic groups have different characteristics regarding the morbidity of GC. The relevance ratio of GC in Uygur is 12.76% (351/2,751) and in Han people is 3.85% (92/2,568). Thus the mortality risk of Uygur people is 2.4 times higher than that of Han people (2,3).
Early symptoms of GC are not obvious. In most GC patients, the disease has progressed to middle or advanced stages when they were diagnosed. However, the prognosis of GC is closely associated with the TNM stage. The 5-year survival rates of GC patients are 90%, 50–60%, and 10–15% for GC stages I, II, and III, respectively (4). Thus, it is important to identify a diagnostic marker of GC which will help early diagnosis and thus prolong the life of GC patients. Serum detection is a convenient and easy technique for disease diagnosis, which is also simple and relatively painless for patients. Thus, identification of serum tumor biomarkers is significant for GC patients.
At present, serum carbohydrate antigen 72-4 (CA72-4), carcinoembryonic antigen (CEA), and carbohydrate antigen 19-9 (CA19-9), are used as tumor markers for GC diagnosis (5). However, their sensitivity and specificity are limited. Long non-coding RNAs (lncRNAs) are greater than 200 nucleotides in length. They can regulate gene expression at both the transcriptional and post-transcriptional levels (6). Thus they play a significant role in the fundamental biological processes of cells and are emerging as new players in the tumorigenic process (7). LncRNAs exhibit specific expression in tissues and can be detected easily in body fluids. This advantage makes them ideal as biomarkers (8), and some, such as H19 (9,10), GAS5 (11), and HOTAIRM1 (12,13), have recently attracted significant attention for the early diagnosis of cancer.
Plasmacytoma variant translocation 1 (PVT1), a new lncRNA, was found to be over expressed in GC tissues and could be a potential biomarker for diagnosis of GC (14,15). However, the expression level of PVT1 in serum is unclear, and whether there are differences between Han and Uygur GC patients is also not known.
In the present study, we investigated and compared the expression level of PVT1 in the serum of Han and Uygur GC patients, and explored the relationship between the expression of PVT1 and clinicopathological features. We analyzed the correlation of PVT1 and AFP, CEA, CA19-9, and CA72-4, in an attempt to reveal the clinical value of PVT1 as a serum biomarker for diagnosis of GC in Han and Uygur patients.
Methods
Sample collection
We collected serum samples from 87 GC patients, comprising 51 Han and 36 Uygur patients, and from 95 normal persons, comprising 55 Han and 40 Uygur. The samples were collected at the First Affiliated Hospital of Shihezi, Xinjiang, China and the First People Hospital of Kashi, Xinjiang, China. Whole venous blood samples (10 mL) were incubated for 30 minutes at room temperature, and centrifuged at low speed (3,000 rpm, 5 min). The supernatant was transferred to new tubes and centrifuged at high speed (12,000 rpm, 10 min) at 4 °C, then the supernatant serum was collected and stored at −80 °C. Clinical data for GC patients and normal controls were obtained by medical record review, from patient records and information, which was anonymized and de-identified prior to analysis. Details of the investigation and the required informed consent were examined and certified by the Ethics Committee of the First Affiliated Hospital School of Medicine, Shihezi University (No. 2016-035-01).
Serum RNA extraction and cDNA synthesis
Total mRNA from serum of GC patients and normal controls was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). A 750 µL aliquot of TRIzol Reagent was added directly to 250 µL of serum, then the RNA was extracted following the manufacturer’s instructions. At the same time, 1.5 mL of fresh blood was collected from each patient, then added to five volumes of 1× erythrocyte lysis buffer, centrifuged at 12,000 rpm for 10 min, and the supernatant was discarded. Two volumes of 1× erythrocyte lysis buffer were added, centrifugation was repeated, and the supernatant was discarded. TRIzol was then used to obtain total RNA from respective blood samples. The integrity of RNA was checked using 1.2% agarose gel electrophoresis (Figure S1A). The purity and concentration of RNA were assessed by measuring the absorbance at 260 and 280 nm using a spectrophotometer. Total RNA from all samples was synthesized using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol and 1.0 µg of RNA was taken from each sample for cDNA synthesis.
Real-time polymerase chain reaction (PCR)
The PVT1 expression level was quantified using a SYBR Green PCR kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol, with the following primers: forward, 5'-GGAAGGTGGAGCGTAAGGA-3' and reverse, 5'-CAATGCCGCCAATCTTGTA-3'. The length of the quantitative PCR product was 92 base pairs. The expression level of PVT1 in each serum sample was normalized to the respective β-actin expression level from blood total RNA, using the following primers: forward, 5'-CCCAGCACAATGAAGATCAAGATCAT-3', and reverse, 5'-ATCTGCTGGAAGGTGGACAGCGA-3' (product length, 101 base pairs). The amplification protocol included an initial heat activation step at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30 s and combined annealing/extension at 55 °C for 30 s. The expression of PVT1 in serum was calculated using the 2(−ΔCT) value and the specificity of each PCR reaction was confirmed by melting curve analysis (Figure S1B,C,D).
Electrochemiluminescence
Samples of venous serum (4 mL) were collected in the morning after overnight fast, and centrifuged at 3,000 rpm for 10 min. The content of tumor markers in serum was detected using AFP, CEA, CA19-9, or CA72-4 specific kits (Roche Diagnostics GmbH Production, Basel, Switzerland) with an automatic immunology analyzer (Roche E170). The tumor markers were defined as positive when above the normal range, and defined as negative when within the range.
Statistical analysis
The rank sum test was used to compare differences between GC patients and normal controls. The correlation between PVT1 and clinicopathological characteristics or tumor markers of GC patients was analyzed by χ2 and Spearman’s test. All statistical analyses were performed using Statistical Package for the Social Sciences software (SPSS, version 20.0, IBM SPSS Statistics for Windows, version 19.0. Armonk, NY, USA). Values of P<0.05 were considered statistically significant.
Results
Serum PVT1 levels were higher in GC patients than in normal controls and PVT1 levels in the serum of Uygur GC patients were higher than those of Han
PVT1 expression levels in serum of 51 Han and 36 Uygur GC patients, together with 55 Han and 40 Uygur normal controls were examined by real-time PCR. According to the rank sum test, PVT1 expression in GC serum was significantly higher than that of normal controls in both Han and Uygur ethnic groups (P<0.05) (Figure 1A,B). However, compared with Uygur GC patients, PVT1 expression level of Han GC patients was significantly lower (P<0.05) (Figure 1C).
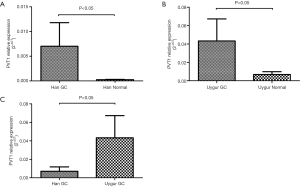
High serum PVT1 level was associated with lymph node metastasis in both Han and Uygur GC patients
Integrated clinical information was available for 28 Han and 31 Uygur GC patients (Table S1). The relationship between serum PVT1 expression level and clinicopathological features was examined in these patients. First, the Han and Uygur GC serum samples were divided into a high PVT1 expression group and a low PVT1 expression group according to the median. The associations between PVT1 expression levels and clinicopathological features of the patients are summarized in Tables 1,2. A significant relationship was found between PVT1 expression and lymph node metastasis in both Han and Uygur patients (P<0.05). However, PVT1 expression level showed no correlation with age, sex, or primary tumor site. Analysis showed that Uygur GC patients were more likely to develop distant metastasis (P<0.05) and more likely to be diagnosed at a late clinical stage (P<0.05) (Table 3).
Table 1
| Variable | Category | PVT1 | P | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Age (year) | 55.29±10.908 | 59.50±11.621 | 0.311 | |
| Sex | Male | 12 | 8 | 0.209 |
| Female | 2 | 6 | ||
| T | T1–2 | 3 | 4 | 1.000 |
| T3–4 | 11 | 10 | ||
| N | N0 | 2 | 8 | 0.046* |
| N1–3 | 12 | 6 | ||
| M | M0 | 13 | 14 | 1.000 |
| M1 | 1 | 0 | ||
| Stage | I, II | 3 | 6 | 0.420 |
| III, IV | 11 | 8 | ||
| Histopathological grade | Poorly | 10 | 7 | 0.440 |
| Well + moderately | 4 | 7 | ||
Note: Fisher’s exact probability test. T, the primary tumor site; N, the involvement of regional lymph node; M, the presence of distant metastatic; *, P<0.05.
Table 2
| Variable | Category | PVT1 | P | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Age (year) | 51.38±14.245 | 59.80±10.936 | 0.078 | |
| Gender | Male | 11 | 11 | 1.000 |
| Female | 5 | 4 | ||
| T | T1–2 | 1 | 1 | 1.000 |
| T3–4 | 15 | 14 | ||
| N | N0 | 0 | 4 | 0.043* |
| N1–3 | 16 | 11 | ||
| M | M0 | 8 | 11 | 0.273 |
| M1 | 8 | 4 | ||
| Stage | I, II | 2 | 2 | 1.000 |
| III, IV | 14 | 13 | ||
Note: Fisher’s exact probability test; T, the primary tumor site; N, the involvement of regional lymph node; M, the presence of distant metastatic; *, P<0.05.
Table 3
| Variable | Category | Han | Uygur | P |
|---|---|---|---|---|
| Age (year) | 57.39±11.266 | 55.45±13.251 | 0.471 | |
| Gender | Male | 20 | 22 | 1.000 |
| Female | 8 | 9 | ||
| T | T1–2 | 7 | 2 | 0.071 |
| T3–4 | 21 | 29 | ||
| N | N0 | 10 | 4 | 0.065 |
| N1–3 | 18 | 27 | ||
| M | M0 | 27 | 19 | 0.001* |
| M1 | 1 | 12 | ||
| Stage | I, II | 9 | 2 | 0.018* |
| III, IV | 19 | 29 |
Note: Fisher’s exact probability test. T, the primary tumor site; N, the involvement of regional lymph node; M, the presence of distant metastatic; *, P<0.05.
Serum PVT1 expression level was correlated with serum CA19-9 level in Han GC patients
Electrochemiluminescence was used to measure serum levels of the tumor markers AFP, CEA, CA19-9, and CA72-4 and the correlation with serum PVT1 level was analyzed in Han and Uygur GC patients by Spearman’s relative analysis. A significant association was found between PVT1expression and CA19-9 in Han GC patients (P<0.05). However, no correlation was found with other tumor markers in Han GC patients (Table 4). In addition, in Uygur people, PVT1 expression levels showed no relationship with any of the four tumor markers (Table 5).
Table 4
| Tumor markers | n | PVT1 | |
|---|---|---|---|
| r | P | ||
| AFP | 51 | 0.013 | 0.926 |
| CEA | 51 | −0.060 | 0.678 |
| CA19-9 | 51 | 0.429 | 0.002* |
| CA72-4 | 51 | 0.000 | 1.000 |
Note: Spearman correlation analysis; r, correlation coefficient. *, P<0.05. PVT1, plasmacytoma variant translocation 1; GC, gastric cancer; AFP, alpha fetoprotein; CEA, carcinoembryonic antigen; CA, carbohydrate antigen.
Table 5
| Tumor markers | n | PVT1 | |
|---|---|---|---|
| r | P | ||
| AFP | 25 | 0.045 | 0.829 |
| CEA | 25 | −0.073 | 0.729 |
| CA19-9 | 25 | −0.097 | 0.643 |
| CA72-4 | 25 | −0.095 | 0.651 |
Note: Spearman correlation analysis; r, correlation coefficient. PVT1, plasmacytoma variant translocation 1; GC, gastric cancer; AFP, alpha fetoprotein; CEA, carcinoembryonic antigen; CA, carbohydrate antigen.
Discussion
GC is a major cause of cancer-related mortality in China (16). Host genetics, bacterial virulence, environmental, and many other factors have all been implicated in affecting the gastric oncogenic process, but the underlying molecular mechanism remains poorly understood.
Han and Uygur people have different genetic backgrounds and their characteristics of morbidity and mortality in GC are also different. For Han and Uygur GC cases, the histopathological features maybe similar, but whether the diagnostic parameters, especially some genetic and molecular biology biomarkers, are suitable for both of these ethnic groups is not known.
In recent years, the results of transcriptomics have indicated that only approximately 2% of genes making up the human genome are protein coding genes. The remaining 98% are transcribed into non-coding RNAs (ncRNAs). Among the non-coding RNAs, 80% are lncRNAs. Interestingly, these lncRNAs could be important biomarkers for clinical diagnosis as well as drug targets for cancer.
Cao et al. (14) found that the lncRNA PVT1 is upregulated in the tumor tissues of GC based on highly significant microarray results. Kong et al. (17) further confirmed that PVT1 is upregulated in GC tumor tissues, and showed that PVT1 silencing can block the G1 phase of the cell cycle, thus halting the proliferation of GC cells. Zhang et al. (18) found that PVT1 promotes the multidrug resistance of GC cells. In addition, PVT1 is located in chromosome 8q24 (19), exactly upstream of the oncogene MYC. All of these results indicate that PVT1 is closely related to the mechanism and treatment of GC.
In addition to the tissues, abnormal expression of lncRNAs can also be detected in body fluid samples, such as serum and saliva (20,21). The expression level of PVT1 in serum is unclear, and whether or not there are difference between Han and Uygur GC patients is also not known. In this study, we collected samples of serum from Han and Uygur GC patients and normal controls. We analyzed the serum expression level of PVT1 and found that the PVT1 expression level was higher in GC patients than in normal individuals both in the Han and Uygur populations. Furthermore, when comparing the Han and Uygur GC patients, we found that the serum PVT1 level in Uygur GC patients was higher than that in Han GC patients. The result of the relationship analysis between serum expression of PVT1 and clinical characteristics suggested that PVT1 could be a marker to identify patients with a tendency for lymphatic metastasis. In a clinical test, if GC patients were found to have a high serum level of PVT1, they would need to be vigilant for the incidence of lymphatic metastasis.
Changes of tumor markers in serum are likely to appear earlier than clinical symptoms. Thus, combined detection of a multiterm tumor marker will be effective in evaluating both the diagnosis and prognosis of GC patients. In one study, patients with an elevated CA19-9 level in serum were found to have more nodal metastases in intrahepatic cholangiocarcinoma (22). Zhou et al. (23) found that CA199 ≥14.06 U/mL and CA125 ≥14.30 U/mL were predictors of endometrial carcinogenesis when entered into the risk mode. The variation of CEA and CA19-9 levels in serum can accurately predict the efficacy of first-line chemotherapy in advanced GC (24). In our research, serum expression levels of PVT1 and CA19-9 showed a correlation in Han GC patients. PVT1 and CA19-9 can thus be combined to diagnose Han GC patients. However, in Uygur GC patients, PVT1 serum expression showed no correlation with tumor markers in serum.
In conclusion, our results suggested that an increase in the serum PVT1 level could be an ideal tumor biomarker for GC diagnosis both in Han and Uygur GC patients. PVT1 level in serum can help to judge the tendency of lymphatic metastasis in GC patients. PVT1 and CA19-9 can be combined as serum tumor markers in Han GC patients. However, whether PVT1 and tumor markers can be combined in Uygur GC patients still needs to be explored. In our future research, we will study the mechanism underlying the high PVT1 level in the serum and the function of PVT1 in GC cells. We plan to analyze the PVT1-protein interaction networks in an attempt to identify the transcription factors or polymerases that are involved in the mechanism of PVT1 in GC cells. We will also test the tumorigenic capacity of PVT1 in gastric cells.
Table S1
| No. | Pathological No. | Age | Gender | Nationality | TNM |
|---|---|---|---|---|---|
| 1 | C8 | 47 | Male | Han | III |
| 2 | C40 | 48 | Male | Han | III |
| 3 | C75 | 70 | Male | Han | IV |
| 4 | C120 | 63 | Male | Han | III–IV |
| 5 | C124 | 63 | Male | Han | III |
| 6 | C127 | 40 | Male | Han | IV |
| 7 | C57 | 58 | Male | Han | II |
| 8 | C95 | 45 | Male | Han | I |
| 9 | C122 | 61 | Male | Han | III |
| 10 | C129 | 55 | Male | Han | II |
| 11 | C135 | 65 | Male | Han | III |
| 12 | C138 | 73 | Male | Han | III |
| 13 | C34 | 43 | Female | Han | III |
| 14 | C60 | 58 | Female | Han | III |
| 15 | C47 | 45 | Female | Han | I |
| 16 | C55 | 43 | Female | Han | III |
| 17 | C72 | 54 | Male | Han | III |
| 18 | C73 | 63 | Male | Han | III |
| 19 | C80 | 77 | Female | Han | III |
| 20 | C81 | 65 | Male | Han | III |
| 21 | C82 | 44 | Female | Han | III |
| 22 | C99 | 64 | Female | Han | II |
| 23 | C125 | 47 | Female | Han | II |
| 24 | C128 | 73 | Male | Han | I |
| 25 | C131 | 78 | Male | Han | III |
| 26 | C137 | 55 | Male | Han | IV |
| 27 | C143 | 46 | Male | Han | I |
| 28 | C104 | 64 | Male | Han | I |
| 29 | K1 | 66 | Male | Uygur | IV |
| 30 | K47 | 49 | Female | Uygur | IV |
| 31 | K13 | 67 | Female | Uygur | IV |
| 32 | K28 | 38 | Male | Uygur | IV |
| 33 | K81 | 56 | Male | Uygur | II |
| 34 | K45 | 40 | Female | Uygur | IV |
| 35 | K49 | 64 | Male | Uygur | IV |
| 36 | K50 | 57 | Male | Uygur | IV |
| 37 | K52 | 83 | Male | Uygur | IV |
| 38 | K55 | 76 | Male | Uygur | IV |
| 39 | K56 | 42 | Female | Uygur | II |
| 40 | K69 | 41 | Male | Uygur | IV |
| 41 | K78 | 40 | Male | Uygur | IV |
| 42 | K84 | 43 | Male | Uygur | IV |
| 43 | K19 | 42 | Female | Uygur | IV |
| 44 | K21 | 40 | Male | Uygur | IV |
| 45 | K3 | 64 | Male | Uygur | IV |
| 46 | K5 | 72 | Male | Uygur | IV |
| 47 | K29 | 68 | Male | Uygur | IV |
| 48 | K68 | 71 | Male | Uygur | IV |
| 49 | K36 | 55 | Female | Uygur | IV |
| 50 | K44 | 50 | Female | Uygur | IV |
| 51 | K61 | 58 | Female | Uygur | IV |
| 52 | K63 | 52 | Male | Uygur | IV |
| 53 | K64 | 39 | Male | Uygur | IV |
| 54 | K76 | 40 | Female | Uygur | IV |
| 55 | K8 | 56 | Male | Uygur | IV |
| 56 | K60 | 72 | Male | Uygur | IV |
| 57 | K65 | 66 | Male | Uygur | III |
| 58 | K72 | 72 | Male | Uygur | II |
| 59 | K74 | 57 | Male | Uygur | II |
GC, gastric cancer.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.12.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital School of Medicine, Shihezi University (No. 2016-035-01), and written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Sun LP, Gong YH, Wang L, et al. Follow-up study on a high risk population of gastric cancer in north China by serum pepsinogen assay. J Dig Dis 2008;9:20-6. [Crossref] [PubMed]
- Ferrari F, Reis MA. Study of risk factors for gastric cancer by populational databases analysis. World J Gastroenterol 2013;19:9383-91. [Crossref] [PubMed]
- Ellison LF, Bryant H, Lockwood G, et al. Conditional survival analyses across cancer sites. Health Rep 2011;22:21-5. [PubMed]
- Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int J Clin Exp Pathol 2015;8:12936-42. [PubMed]
- Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer 2016;15:43. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer 2016;15:39. [Crossref] [PubMed]
- Hashad D, Elbanna A, Ibrahim A, et al. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. J Clin Lab Anal 2016;30:1100-5. [Crossref] [PubMed]
- Chen JS, Wang YF, Zhang XQ, et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma 2016;63:223-30. [PubMed]
- Han L, Ma P, Liu SM, et al. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol 2016;37:6847-54. [Crossref] [PubMed]
- Wan L, Kong J, Tang J, et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med 2016;20:2036-44. [Crossref] [PubMed]
- Heubach J, Monsior J, Deenen R, et al. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol Cancer 2015;14:108. [Crossref] [PubMed]
- Cao WJ, Wu HL, He BS, et al. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol 2013;19:3658-64. [Crossref] [PubMed]
- Yuan CL, Li H, Zhu L, et al. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma 2016;63:442-9. [Crossref] [PubMed]
- Zhou DD, Liu XF, Lu CW, et al. Long non-coding RNA PVT1: Emerging biomarker in digestive system cancer. Cell Prolif 2017;50: [Crossref] [PubMed]
- Kong R, Zhang EB, Yin DD, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer 2015;14:82. [Crossref] [PubMed]
- Zhang XW, Bu P, Liu L, et al. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun 2015;462:227-32. [Crossref] [PubMed]
- Colombo T, Farina L, Macino G, et al. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int 2015;2015:304208. [Crossref] [PubMed]
- Xie H, Ma H, Zhou D. Plasma HULC as a Promising Novel Biomarker for the Detection of Hepatocellular Carcinoma. Biomed Res Int 2013;2013:1-5. [PubMed]
- Majem B, Rigau M, Reventós J, et al. Non-Coding RNAs in Saliva: Emerging Biomarkers for Molecular Diagnostics. Int J Mol Sci 2015;16:8676-98. [Crossref] [PubMed]
- Bergquist JR, Ivanics T, Storlie CB, et al. Implications of CA19-9 elevation for survival, staging, and treatment sequencing in intrahepatic cholangiocarcinoma: A national cohort analysis. J Surg Oncol 2016;114:475-82. [Crossref] [PubMed]
- Zhou L, Meng Z, Wu Y, et al. Prediction of endometrial carcinogenesis probability while diagnosed as atypical endometrial hyperplasia: a new risk model based on age, CA199 and CA125 assay. Eur J Obstet Gynecol Reprod Biol 2014;183:5-9. [Crossref] [PubMed]
- He B, Zhang HQ, Xiong SP, et al. Changing patterns of Serum CEA and CA199 for Evaluating the Response to First-line Chemotherapy in Patients with Advanced Gastric Adenocarcinoma. Asian Pac J Cancer Prev 2015;16:3111-6. [Crossref] [PubMed]


