
GEFT aberrant expression in soft tissue sarcomas
Introduction
Soft tissue sarcoma (STS) is highly heterogenous and contributes to 10% of all childhood tumors (1,2). According to the classification of the World Health Organization (WHO), STS has more than 60 subtypes that are prone to form pseudocapsule and mistaken as benign (3). Therefore, identifying novel biomarkers of STS is crucial for early diagnosis and treatment. STS may occur in all non-epithelial tissues, including muscle, bone, lymphatic, hematopoietic, and glial tissues. Based on the molecular genetics, these STSs generated from different tissues are categorized as chromosomal translocation-associated sarcomas (CTAS) and non-chromosomal translocation-associated sarcomas (NCTAS) (4).
Generally, CTAS involves specific genetic alterations with simple karyotypes, including chromosomal translocations and gene fusions generated, such as alveolar rhabdomyosarcoma, Ewing’s sarcoma, and myxoid liposarcoma (4). By contrast, NCTAS lacks specific genetic change and exhibits complex karyotypes, such as leiomyosarcoma, pleomorphic rhabdomyosarcoma, and undifferentiated sarcoma (4). These two subgroups of STS exhibit different gene expression profiles, histological morphologies, and karyotypes. Moreover, distinctions in tumor tumorigenesis, development, and prognosis may be apparent. Therefore, discovering new molecular markers, which can accurately distinguish the subgroups of STS for diagnosis, treatment, and prognosis assessment, is important.
Guanine nucleotide exchange factor T (GEFT) is a member of the guanine nucleotide exchange factor (GEF) family and is located on chromosome 12q13.3 (5). GEFT up-regulate activity of Rho GTPases such as Rac1, and Cdc42, RhoA, by stimulating the exchange of an inactive GDP-bound state and an active GTP-bound state, which activates upstream receptor signaling (6-8). Ongherth et al. (9) have reported that GEFT involved in secretion in cardiac fibroblasts and influences cardiac remodeling. GEFT can enhance dendritic spine formation and neurite outgrowth in neuroblastoma cells by activating the Rac/Cdc42-PAK signaling pathway (10). GEFT plays a crucial role in breast cancer progression and metastasis (11,12). Furthermore, other members of the Rho GEF family can modulate various functions through targeting Rho GTPases (13) that are involved in tumorigenesis, invasion and metastasis of tumors (14-17).
High copy numbers of GEFT is identified by high-resolution array comparative genomic hybridization in rhabdomyosarcoma tissues (18). Moreover, the rate of GEFT protein expression is remarkably up-regulated in rhabdomyosarcoma tissues, the level of its expression is related to lymph node and distant metastasis, advanced disease stages (stages III/IV), and poor overall survival (19). In this study, we identified if GEFT also exhibits abnormal expression in various subtypes of STS.
In the present study, GEFT protein expression was detected in multiple subtypes of STS and normal striated muscle tissues and compared the differences in clinical pathology, histological feature, and expression levels of GEFT protein and mRNA between CTAS and NCTAS. The rates of GEFT positive expression and overexpression were higher in multiple subtypes of STS than in the controls. The differences between the levels of GEFT protein and mRNA expression were not statistically significant between CTAS and NCTAS. GEFT may play an important role in tumorigenesis of STS.
Methods
Patient samples
A total of 219 FFPE STS samples (43 leiomyosarcoma, 38 dermatofibrosarcoma protuberans, 36 undifferentiated sarcoma, 21 liposarcoma, 18 fibrosarcoma, 18 Ewing’s sarcoma, and 45 rhabdomyosarcoma) and their clinical data were collected from the department of pathology of the First Affiliated Hospital of Shihezi University School of Medicine between 1972 and 2015. The samples were divided into two major types based on molecular genetic studies. A total of 36 normal striated muscle tissues as control were available. All hematoxylin and eosin (H&E) stained sections underwent histological examination by two or more highly qualified pathologists, who also considered the patient age, location of tumor, tumor size, and the results of immunohistochemical staining for diagnosis and differential diagnosis. The results were reported in accordance with the World Health Organization classification of soft tissue and osteosarcoma fourth edition [2013]. The study was recognized by the institutional ethics committee at the First Affiliated Hospital of Shihezi University School of Medicine (Approval ID: 2015-076-01) and strictly conducted in accordance with the ethical guidelines of the Declaration of Helsinki.
Clinical pathology and histological study
Pathomorphological observation was mainly based on the volume, size, and shape of the tumor cells. All case studies were divided into three categories, including small round, spindle, and pleomorphic cell tumors.
Tissue microarray (TMA) construction
Samples of each paraffin-embedded tissue were sectioned and stained with H&E. On the screened HE sections, typical regions of STS were marked and the corresponding area on the wax block was searched as the target for TMA construction. Minicore instrument and TMS Designer2 software were utilized in the design and creation of tissue cores. A hollow needle with a diameter of 1.0 mm was used to collect the tissue blocks and inserted into a blank paraffin block with the tissue array. Each typical region was reviewed using the hollow needle to ensure that at least 70% of the samples can be unfolded.
Immunohistochemistry
GEFT expression was detected by subjecting paraffin-embedded tissue sections to standard immunohistological staining techniques using the Envision system (Dako, Carpinteria, CA). The 3.5 µm-thick sections were prepared and incubated at 65 °C for 4–6 h and then deparaffinized by 3 consecutive treatments with xylene and alcohol. Endogenous peroxidase activity was blocked with 3% H2O2 for 10 min. Antigen sites were unmasked by heating the sections in a microwave oven under 750 W for approximately 8 min in a medlar yoghurt buffer. The slices were washed thrice with PBS for 5 min and blocked by treatment with anti-GEFT antibody (Abcam, Cambridge, USA; dilution 1:300) at 4 °C overnight. The EnVision kits (Abcam, Cambridge, USA) was used as the second antibody in accordance with experiment requirements and incubated for 30 min. The sections were stained with -diaminobenzidine, counterstained with Mayer’s hematoxylin, and dehydrated with graded alcohol and xylene. Finally, the slides were cleared and mounted. Semi-quantitative analysis of GEFT expression was performed using the sections obtained. Samples were defined as GEFT-positive when brownish-yellow particles in the nucleus were present. Signal intensity was scored as 0, 1–3, 4–6, or 7–9 for no staining, weak staining, medium staining, and strong staining, respectively. The total score was calculated by multiplying the score of positive cell range by nuclear staining intensity score. Nuclear staining intensity was scored as 0–3, which corresponded to no staining, buff, yellow, and brown staining, respectively. Similar to the nuclear staining intensity scores, the percentage of positive cell range was scored as 0, 1, 2 or 3 for ≤5% of the cells range, 6–35% of the cell range, 36–65% of the cell range or ≥66% of the cells range, respectively. Two experienced pathologists reviewed the immunostaining results by randomly selecting 5 regions from each sample section using a 400× power microscope.
Quantitative real-time polymerase chain reaction (qRT-PCR) detection
Expression of the GEFT mRNA in STS was detected using qRT-PCR. RNA was extracted from tumor samples according to the specifications of the manufacturers using RNeasy FFPE Kit (Qiagen, Hilden, Germany). The mRNA was transcribed into single-stranded cDNA by reverse transcription using QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). Quantitative RT-PCR was performed through SYBR green PCR Master Mix (Qiagen, Hilden, Germany) with the Applied Biosystems 7500. All PCR assays were performed in triplicates. Triplicate Ct values were averaged, and the relative expression levels of GEFT were determined as 2−ΔCt.
Statistical analysis
The Statistical Package for the Social Science (SPSS) (IBM Corp., Armonk, NY, USA) version 17.0 was used for all statistical analyses. Correlations between GEFT staining were calculated using the χ2 tests. Two-sample t-tests were conducted to compare GEFT mRNA expression between CTAS and NCTAS. P-values were calculated by the Epi-Info program, and P value of less than 0.05 were considered significant.
Results
Clinical and pathological characteristics
The clinicopathological data of all tumor samples are listed in Table 1. The mean age of all the patients was 39.72 years. CTAS occurred in 89 cases with younger age range (mean 34.22 years) compared with that of NCTAS cases, which occurred in 130 cases at an older age range (mean 45.22 years). Histopathological observation showed that CTAS was mostly composed of small round and spindle cells, whereas NCTAS comprised pleomorphic cells.
Table 1
| Organization type | Case (N) | Average age (year) | Male/female ratio |
|---|---|---|---|
| CTASs | 89 | 35.62 | 1:1.2 |
| Alveolar rhabdomyosarcoma | 20 | 20.40 | 1:1 |
| Ewing’s sarcomas | 18 | 26.21 | 1.5:1 |
| Dermatofibrosarcoma protuberans | 38 | 44.70 | 1:1.3 |
| Myxoid liposarcoma | 13 | 45.59 | 1:3 |
| NCTASs | 130 | 43.51 | 1:1 |
| Embryonal/pleomorphic rhabdomyosarcoma | 25 | 20.40 | 1:1 |
| Dedifferentiated liposarcoma | 8 | 59.29 | 1:1 |
| Leiomyosarcoma | 43 | 47.65 | 1:2.3 |
| Undifferentiated sarcoma | 36 | 46.96 | 2.3:1 |
| Adult fibrosarcoma | 18 | 51.78 | 2.3:1 |
| Total | 219 | 40.30 | 1:1.1 |
CTAS, chromosomal translocation-associated sarcoma; NCTAS, non-chromosomal translocation-associated sarcoma.
GEFT expression in STS
The results showed that the overall GEFT protein expression and overexpression rates were 89.50% (196/219) and 51.60% (113/219) in the 219 STS cases, respectively. The rate of 36 normal striated muscle control samples expressing GEFT (1+) were 13.89% (5/36). Statistically significant difference was observed between the rate of positive GEFT expression and the controls (χ2=105.89, P<0.05). Moreover, the overall percentages of CTAS and NCTAS samples expressing GEFT were 89.89% (80/89) and 80.99% (116/130), respectively. The percentages of samples overexpressing GEFT (categorized as 2+ or 3+ staining) were 48.31% (43/89) and 53.85% (70/130) for CTAS and NCTAS, respectively. The positive expression (χ2=0.02, P>0.05) and overexpression rates (χ2=0.65, P>0.05) between CTAS and NCTAS did not exhibit any statistically significant difference. The rates of GEFT protein expression in CTAS and NCTAS are shown in Table 2.
Table 2
| Organization type | Case (N) | GEFT | |||
|---|---|---|---|---|---|
| – (%) | 1+ (%) | 2+ (%) | 3+ (%) | ||
| CTASs | 89 | ||||
| Alveolar rhabdomyosarcoma | 20 | 0 (0.00) | 4 (20.00) | 8 (40.00) | 8 (40.00) |
| Ewing’s sarcomas | 18 | 0 (0.00) | 6 (33.33) | 6 (33.33) | 6 (33.33) |
| Dermatofibrosarcoma protuberans | 38 | 4 (10.53) | 22 (57.89) | 12 (31.58) | 0 (0.00) |
| Myxoid liposarcoma | 13 | 5 (38.46) | 5 (38.46) | 3 (23.08) | 0 (0.00) |
| NCTASs | 130 | ||||
| Embryonal/pleomorphic rhabdomyosarcoma | 25 | 3 (12.00) | 7 (28.00) | 8 (32.00) | 7 (28.00) |
| Dedifferentiated liposarcoma | 8 | 2 (25.00) | 4 (50.00) | 2 (25.00) | 0 (0.00) |
| Leiomyosarcoma | 43 | 9 (20.93) | 20 (46.51) | 13 (30.23) | 1 (2.32) |
| Undifferentiated sarcoma | 36 | 0 (0.00) | 7 (19.44) | 18 (50.00) | 11 (30.56) |
| Adult fibrosarcoma | 18 | 0 (0.00) | 8 (44.44) | 10 (55.56) | 0 (0.00) |
| Total | 219 | 23 (10.50) | 83 (37.90) | 80 (36.53) | 33 (15.07) |
GEFT, guanine nucleotide exchange factor T; CTAS, chromosomal translocation-associated sarcoma; NCTAS, non-chromosomal translocation-associated sarcoma.
GEFT expression in CTAS
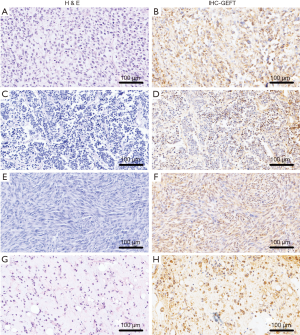
GEFT protein expression in alveolar rhabdomyosarcoma
The rate of GEFT expression in alveolar rhabdomyosarcoma was 100% (20/20), including 8, 8, and 4 cases of strongly positive (3+), positive (2+), and weakly positive (+) expressions, respectively. GEFT overexpression (2+/3+) occurred in 80% (16/20) of cases. The positive expression and overexpression rates were 13.89% (5/36) and 0.00% (0/36) in the control samples, respectively. The rates of GEFT expression (χ2=38.57, P<0.05) and overexpression (χ2=40.32, P<0.05) in alveolar rhabdomyosarcoma were more significant than in the controls.
GEFT protein expression in Ewing’s sarcoma
All 18 Ewing’s sarcoma samples were positive for GEFT protein expression (18/18), including 6 cases each of strongly positive (3+), positive (2+), and weakly positive (+) expression. GEFT overexpression (2+/3+) occurred in 66.67% (12/18) of samples. The rates of GEFT expression (χ2=36.39, P<0.05) and overexpression (χ2=30.86, P<0.05) in Ewing’s sarcoma were higher than in the controls.
GEFT protein expression in dermatofibrosarcoma protuberans
The rate of positive GEFT protein expression was 89.5% (34/38) in dermatofibrosarcoma protuberans, whereas that of the overexpression (2+/3+) was 31.58% (12/38). The positive (χ2=42.37, P<0.05) and overexpression rate differences (χ2=13.57, P<0.05) were statistically significant compared with those in the control group.
GEFT protein expression in myxoid liposarcoma
The percentages of samples expressing GEFT in experimental group were 61.54% (8/13) for myxoid liposarcoma, including positive (2+), weakly positive (1+), and GEFT overexpression (2+) in 3, 5, and 23.08% (3/13) cases, respectively. The rate of positive GEFT expression (χ2=14.58, P<0.05) and overexpression (χ2=8.85, P<0.05) in myxoid liposarcoma were significantly higher compared with those in the controls.
The rates of GEFT expression and overexpression in CTAS were 89.89% (80/89) and 48.31% (43/89), respectively. Statistically significant difference was observed between the rate of GEFT positive expression (χ2=68.03, P<0.05) and overexpression (χ2=26.51, P<0.05) in 93 CTAS cases compared with 36 normal controls. The results of CTAS are shown in Figure 1.
GEFT protein expression in NCTAS
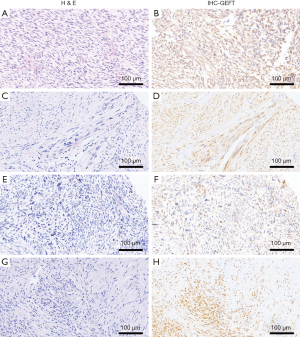
GEFT protein expression in embryonal and pleomorphic rhabdomyosarcoma
The rate of positive GEFT protein expression and overexpression (2+/3+) were 88% (22/25) and 60% (15/25), respectively. Compared with the controls, the positive expression (χ2=32.85, P<0.05) and overexpression rate (χ2=28.64, P<0.05) differences were statistically significant.
GEFT protein expression in dedifferentiated liposarcoma
Two strongly positive and four weakly positive cases were observed in the eight enrolled samples. The rate of GEFT positive expression and overexpression (2+) was 75% (6/8) and 25% (2/8), respectively. The difference between positive expression (χ2=13.04, P<0.05) and overexpression rate (χ2=9.42, P<0.05) was statistically significant compared with that of controls.
GEFT protein expression in leiomyosarcoma
The rate of GEFT expression in the experimental group with leiomyosarcoma was 79.07% (34/43), including strongly positive (3+), positive (2+), and weakly positive (+) in 1, 13, and 20 cases, respectively. GEFT overexpression (2+/3+) occurred in 32.56% (14/43) of cases. The rate of GEFT expression (χ2=33.30, P<0.05) and overexpression (χ2=14.25, P<0.05) in leiomyosarcoma was higher than in the controls.
GEFT protein expression in undifferentiated sarcoma
A total of 36 undifferentiated sarcoma samples positively expressed GEFT protein, including 11, 18, and 7 strongly positive (3+), positive (2+), and weakly positive (+) samples, respectively. Approximately 80.56% (29/36) exhibited GEFT overexpression. The difference between positive expression (χ2=54.44, P<0.05) and overexpression (χ2=48.59, P<0.05) rates was statistically significant compared with the controls.
GEFT protein expression in adult fibrosarcoma
GEFT expression occurred in 100% of (18/18) adult patients with fibrosarcoma, including 10 and 8 cases of positive (2+) and of weakly positive (+) expressions, respectively. GEFT was overexpressed (2+/3+) in 55.56% (10/18) of samples. Compared with the controls, the positive expression (χ2=36.39, P<0.05) and overexpression (χ2=24.55, P<0.05) rate differences were statistically significant.
In general, the overall rates of GEFT expression and overexpression in NCTAS were 80.89% (116/130) and 53.85% (70/130), respectively. Compared with the control group, the positive (χ2=69.58, P<0.05) and overexpression (χ2=33.52, P<0.05) rate differences were statistically significant. The results of NCTAS are shown in Figure 2.
GEFT mRNA expression in STS tissues by qRT-PCR
We analyzed the expression of GEFT mRNA in 62 cases of STS by qRT-PCR. The analysis included 27 CTAS (fusion gene+) and 35 NCTAS specimens (fusion gene−). The differences of GEFT mRNA expression were not statistically significant between CTAS and NCTAS (P>0.05).
Discussion
With the development of molecular genetics, a large number of researchers have discovered chromosomal translocation and fusions genes formation in some types of STS, and these findings are crucial in early diagnosis, accurate identification, and targeted therapies. Based on molecular genetics, STS can be classified as CTAS and NCTAS. The two types of STS display obvious differences in terms of molecular pathogenesis, gene expression profiles, age of patients, and histopathological morphologies.
Given that accurately distinguishing different type of STS is critical for clinical diagnosis, we compared the differences in the age of the patients and histological features between CTAS and NCTAS. The results showed the age range of onset of CTAS patients (mean 34.22 years) was lower than the age range of onset of NCTAS patients (mean 45.22 years). The results are consistent with a study by Borden et al. (20) in which CTAS occurred with younger age range (mean 27 years) compared with that of NCTAS cases, which occurred at an older age range (mean 57 years). Histopathological observation suggested that CTAS was mostly composed of small round and spindle cells, whereas NCTAS comprised pleomorphic cells. These observations were also consistent with those of a previous study conducted by Borden et al. (20). However, distinguishing the type is still difficult when the tumors suggest very little evidence of differentiation. Thus, discovering biomarkers of distinguish STS is especially important for the diagnosis of STS.
GEFT is a critical regulator by cycling between a GDP-bound inactive form and a GTP-bound active form on Rho GTPases for cell processes. Targeting Rho GTPases of GEFT are mainly RhoA, Rac1, and Cdc42 (6,7,21). GEFT induces cell cytoskeleton reorganization and cell morphology change by activating Rac/Cdc42 Rho GTPase (5). In addition, GPR116 can regulate cell motility and morphology through the GEFT-RhoA/Rac1 signaling cascades and involved in breast carcinoma progression and metastasis (11). GEFT plays an important role in a variety of cellular processes, including angiotensin II-mediated cell elongation, cell proliferation, and stress fiber formation (22). Obviously, the cellular functions of GEFT involved in regulation are associated with tumorigenesis and progression.
However, the role of GEFT in STS remains elusive. Previous studies have only revealed the role of GEFT in RMS. Sun et al. (19) have reported increased GEFT amplification in rhabdomyosarcoma. In this study, aberrant expression of GEFT exists in multiple subtypes of STS. Moreover, the rates of GEFT positive expression and overexpression in all subtypes of STS detected were significantly higher than that in normal controls. These findings are consistent with our observation of increasing GEFT expression in RMS. To the best of our knowledge, this study is the first to reveal the expression levels of GEFT in various subtypes of STS. As the gene expression level is correlated with its tumorigenesis (18), GEFT may play an important role in tumorigenesis of STS.
In addition, several studies have reported the differences between CTAS and NCTAS in P53 gene changes, length of telomere, karyotype complexity, and maintenance mechanisms (4,23-25). The formation of fusion genes is more important in a number of differences between CTAS and NCTAS. CTAS may change certain genes and turn into fusion genes with new structures and oncogenic functions. Stoll et al. (26) have revealed that silencing of the EWS-FLI1 fusion gene produced by chromosomal translocation in Ewing’s sarcoma leads to cell cycle changes and induction of apoptosis. The PAX3-FOXO1 fusion gene generated in alveolar rhabdomyosarcoma can promote proliferation, invasion, cell survival, and inhibition of terminal differentiation (27). Moreover, the gene fusion and the environment of cellular may interact with each other, where the environment of cells is involved in the choice of oncogenic fusion, fusion genes generated in turn alter the microenvironment of cells and influence tumorigenesis (28). Fusion-encoded proteins regulate the transcription of genes closely related to certain key functions of cells, such as cell proliferation (29). We further analyzed the difference in GEFT protein expression levels between CTAS and NCTAS. However, no significant difference between CTAS and NCTAS was present. Since fusion proteins often affect transcription factors involved in regulating certain key functions of cells, we detected the levels of GEFT mRNA in 27 CTAS and 35 NCTAS specimens. The differences of GEFT mRNA expression were not statistically significant between CTAS and NCTAS. Our results suggest that GEFT plays a similar role in CTAS and NCTAS. However, these observations are inconsistent with our expected results. The differences between the results may be due to the limitations of the small sample sizes and the diversity of tumor subtypes detected. Further study using increased sample sizes is necessary to clarify the relationship between GEFT expression and chromosome translocation.
In conclusion, the levels of GEFT expression in STS and its role in CTAS and NCTAS were preliminarily described. These results suggest aberrant expression of GEFT exists in multiple subtypes of STS. No difference of GEFT expression was detected between CTAS and NCTAS. GEFT may play an important role in tumorigenesis of STS, which can provide some reference value for clinical diagnosis of STS. This study may lay a foundation for an in-depth understanding of the molecular mechanisms of STS.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was recognized by the institutional ethics committee at the First Affiliated Hospital of Shihezi University School of Medicine (Approval ID: 2015-076-01) and strictly conducted in accordance with the ethical guidelines of the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lauer S, Gardner JM. Soft tissue sarcomas--new approaches to diagnosis and classification. Curr Probl Cancer 2013;37:45-61. [Crossref] [PubMed]
- Doyle LA. Sarcoma classification: an update based on the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone. Cancer 2014;120:1763-74. [Crossref] [PubMed]
- Wendtner CM, Delank S, Eich H. Multimodality therapy concepts for soft tissue sarcomas. Internist (Berl) 2010;51:1388-96. [Crossref] [PubMed]
- Liu C, Li B, Li L, et al. Correlations of telomere length, P53 mutation, and chromosomal translocation in soft tissue sarcomas. Int J Clin Exp Pathol 2015;8:5666-73. [PubMed]
- Guo X, Stafford LJ, Bryan B, et al. A Rac/Cdc42-specific exchange factor, GEFT, induces cell proliferation, transformation, and migration. J Biol Chem 2003;278:13207-15. [Crossref] [PubMed]
- Smith TK, Hager HA, Francis R, et al. Bves directly interacts with GEFT, and controls cell shape and movement through regulation of Rac1/Cdc42 activity. Proc Natl Acad Sci U S A 2008;105:8298-303. [Crossref] [PubMed]
- van Unen J, Reinhard NR, Yin T, et al. Plasma membrane restricted RhoGEF activity is sufficient for RhoA-mediated actin polymerization. Sci Rep 2015;5:14693. [Crossref] [PubMed]
- Souchet M, Portales-Casamar E, Mazurais D, et al. Human p63RhoGEF, a novel RhoA-specific guanine nucleotide exchange factor, is localized in cardiac sarcomere. J Cell Sci 2002;115:629-40. [PubMed]
- Ongherth A, Pasch S, Wuertz CM, et al. p63RhoGEF regulates auto- and paracrine signaling in cardiac fibroblasts. J Mol Cell Cardiol 2015;88:39-54. [Crossref] [PubMed]
- Bryan B, Kumar V, Stafford LJ, et al. GEFT, a Rho family guanine nucleotide exchange factor, regulates neurite outgrowth and dendritic spine formation. J Biol Chem 2004;279:45824-32. [Crossref] [PubMed]
- Tang X, Jin R, Qu G, et al. GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Galphaq-p63RhoGEF-Rho GTPase pathway. Cancer Res 2013;73:6206-18. [Crossref] [PubMed]
- Hayashi A, Hiatari R, Tsuji T, et al. p63RhoGEF-mediated formation of a single polarized lamellipodium is required for chemotactic migration in breast carcinoma cells. FEBS Lett 2013;587:698-705. [Crossref] [PubMed]
- van Unen J, Yin T, Wu YI, et al. Kinetics of recruitment and allosteric activation of ARHGEF25 isoforms by the heterotrimeric G-protein Galphaq. Sci Rep 2016;6:36825. [Crossref] [PubMed]
- Leabu M, Uniyal S, Xie J, et al. Integrin alpha2beta1 modulates EGF stimulation of Rho GTPase-dependent morphological changes in adherent human rhabdomyosarcoma RD cells. J Cell Physiol 2005;202:754-66. [Crossref] [PubMed]
- Poudel KR, Roh-Johnson M, Su A, et al. Competition between TIAM1 and Membranes Balances Endophilin A3 Activity in Cancer Metastasis. Dev cell 2018;45:738-52.e6. [Crossref] [PubMed]
- Haga RB, Ridley AJ. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016;7:207-21. [Crossref] [PubMed]
- Orgaz JL, Herraiz C, Sanz-Moreno V. Rho GTPases modulate malignant transformation of tumor cells. Small GTPases 2014;5:e29019. [Crossref] [PubMed]
- Liu C, Li D, Hu J, et al. Chromosomal and genetic imbalances in Chinese patients with rhabdomyosarcoma detected by high-resolution array comparative genomic hybridization. Int J Clin Exp Pathol 2014;7:690-8. [PubMed]
- Sun C, Liu C, Li S, et al. Overexpression of GEFT, a Rho family guanine nucleotide exchange factor, predicts poor prognosis in patients with rhabdomyosarcoma. Int J Clin Exp Pathol 2014;7:1606-15. [PubMed]
- Borden EC, Baker LH, Bell RS, et al. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res 2003;9:1941-56. [PubMed]
- Momotani K, Somlyo AV. p63RhoGEF: a new switch for G(q)-mediated activation of smooth muscle. Trends Cardiovasc Med 2012;22:122-7. [Crossref] [PubMed]
- Wuertz CM, Lorincz A, Vettel C, et al. p63RhoGEF--a key mediator of angiotensin II-dependent signaling and processes in vascular smooth muscle cells. FASEB J 2010;24:4865-76. [Crossref] [PubMed]
- Pfeifer JD, Hill DA, O'Sullivan MJ, et al. Diagnostic gold standard for soft tissue tumours: morphology or molecular genetics? Histopathology 2000;37:485-500. [Crossref] [PubMed]
- Nilbert M, Meza-Zepeda LA, Francis P, et al. Lessons from genetic profiling in soft tissue sarcomas. Acta Orthop Scand Suppl 2004;75:35-50. [Crossref] [PubMed]
- Ladanyi M, Bridge JA. Contribution of molecular genetic data to the classification of sarcomas. Hum Pathol 2000;31:532-8. [Crossref] [PubMed]
- Stoll G, Surdez D, Tirode F, et al. Systems biology of Ewing sarcoma: a network model of EWS-FLI1 effect on proliferation and apoptosis. Nucleic Acids Res 2013;41:8853-71. [Crossref] [PubMed]
- Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett 2008;270:10-8. [Crossref] [PubMed]
- Barr FG. Translocations, cancer and the puzzle of specificity. Nat Genet 1998;19:121-4. [Crossref] [PubMed]
- de Alava E. Molecular pathology in sarcomas. Clin Transl Oncol 2007;9:130-44. [Crossref] [PubMed]


