The clinical value of circulating tumor DNA detection in advanced non-small cell lung cancer
IntroductionOther Section
With the highest mortality among all cancer-related deaths, lung cancer is always a kind of highly malignant disease that severely threatens human life. Approximately 1.8 million people, 13% of all new cancer cases were diagnosed to be lung cancer in 2012 worldwide (1). The morbidity of lung cancer in China is also belonging to a high incidence rate. According to the latest investigations, there were 0.73 million new patients and 0.61 million deaths due to lung cancer in China every year, and indicating that lung cancer has become the leading killer of malignant tumors in China (2). Lung cancer includes non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) according to its histological and pathological characteristics, and NSCLC accounts for 80% to 85% of all lung cancer. Metastasis in NSCLC patients always represent a poor prognosis, with a median survival of 3 to 6 months (3). To our disappointment, 10–25% of the newly diagnosed advanced NSCLC patients are commonly accompanied with metastasis, and even in those with orthotopic tumor, 20–40% would develop into brain metastasis and 30–40% would develop into bone metastasis in the following treatment (4-7).
The conventional therapy for NSCLC includes operation, chemotherapy, radiotherapy, and targeted therapy, combined treatment and multi-line drugs are chosen to prolong the survival of advanced cohorts. However, there is significant difference appeared in disease progression and drug resistance among NSCLC individuals, which suggesting that it is necessary to recognize the various characteristics of NSCLC from different perspectives, such as molecular marker or DNA marker (8). The circulating tumor DNA (ctDNA), existing in peripheral blood is a kind of DNA fragment comes from tumor cells and carries molecular mutational information (including single nucleotide variant, deletions, insertions, rearrangements, copy number variants, and methylation, etc.) of different cells, such as necrotic, apoptotic tumor cells, circulating tumor cells, and exosomes secreted by tumor cells (9). As a dynamic biomarker of tumor cells, a series of remarkable discoveries indicated ctDNA to be valuable in disease progression and drug resistance in advanced NSCLC (10-12). For example, ctDNA detection revealed that epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) drugs resistance chiefly derived from the T790M mutation (threonine to methionine substitution at codon 790) in EGFR, and the aberrations of MET was associated with poor prognosis (13,14).
In this study, high-throughput sequencing technology and bioinformatic analysis were used to explore mutational spectrum of the ctDNA which was extracted from peripheral blood of patients with advanced NSCLC. We characterized the molecular features of post-treatment patients and analyzed the correlation of genetic mutation and prognosis, and results may serve as the reference of subsequent precision medicine, accurate illness monitoring and prognosis.
MethodsOther Section
Patient cohorts and ethics statement
This subject recruited the cancer patients who underwent ctDNA detection in Zhejiang Cancer Hospital from January 2017 to October 2017, and out of which 36 patients presenting with NSCLC in stage IIIB–IVB (International Union Against Cancer criteria) were eventually enrolled in study.
All procedures in this study had been approved by the Ethics Committee of Zhejiang Cancer Hospital (IRB-2018-201), and the methods were carried out in accordance with the approved guidelines. All patients involved in this research provided written informed consent for the use of blood samples, and the application of clinical medication was under the approval of the Ethics Committee.
Samples collection and DNA extraction
Streck blood collection tubes were used to collect 10 mL peripheral blood of every patient enrolled. The tubes were gently turned upside down for blending at least 10 times, and then were stored at 6–25 °C until use. For surgical tissue samples, the total mass of each sample was required no less than 60 mg, the proportion of tumor cells was required no less than 70%, and the proportion of necrotic cells was required no more than 10%. No less than 3 needles were required for percutaneous puncture samples. All tissue samples were stored in DNA preservation tubes (1Gene, Hangzhou, China) and were handled in 2 days.
DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) was used to extract cell free DNA and genomic DNA from blood samples and tissue samples respectively. Agarose gel electrophoresis was performed to test the degradation and protein/RNA pollution of extracted DNA. Concentration of extracted DNA was determined by Qubit dsDNA HS assay kit on Qubit Fluorometer according to the manufacturing protocol (Life Technologies, Carlsbad, CA, USA). DNA quality (OD 260/280) was measured by Nanodrop (Thermo Fisher Scientific, USA). The extracted DNA sample which had an OD ratio between 1.8 and 2.0 and a mass more than 10 ng was considered qualified and would be used for following library construction.
Library preparation and target-hybrid capturing
Library preparation was performed as the manufacturer’s instructions of KAPA Hyper prep kit (Illumina Co., Ltd, San Diego, USA). In short, the fragmental DNA was respectively subjected to terminal repair, A-tailing adenylation, and ligation to indexed adapters. The targeted sequencing is a custom hybridization capture based assay which was designed by 1Gene, Inc., (Hangzhou, China). After library construction and hybridization capture, the targeted DNA fragments were amplified by polymerase chain reaction.
Sequencing and bioinformatic processing
PE150 sequencing was performed on Illumina CN500 platform. The quality control of raw data was conducted, and the clean reads were obtained after removing adaptor sequences and low mapping quality reads. For variant analysis, sequences were aligned to the reference human genome (hg19) with BWA software. The linkage of sequencing fragments was optimized by GATK, and the annotation of SNP/Indel was acquired with VarScan2. The sequencing coverage of targeted positions was required ≥90%, and the average sequencing depth of ctDNA samples and tDNA samples was respectively required ≥1,000× and ≥500×. A variant with a variant frequency ≥0.3% for ctDNA samples or ≥1.0% for tDNA samples was considered credible. Final results were filtered through 1,000 genomes database and dbSNP database.
Statistical analysis
All data were statistically analyzed by statistical software SPSS 20.0. Measurement data was exhibited as mean ± standard deviation (SD). The relationship between mutational genes and clinicopathology was measured by Fisher’s exact or Chi-square tests as appropriate. Log-rank test was used for survival analysis. Two-sided P<0.05 was considered as statistical significance.
ResultsOther Section
Patient characteristics
The clinicopathological characteristics of enrolled patients are presented in Table 1. This study totally recruited 36 NSCLC patients with median age of 62 years old (range, 46 to 74 years old), including 18 males and 18 females. According to the histological classification of lung cancer published by World Health Organization, 28 patients were diagnosed as adenocarcinoma and 8 were squamous cell carcinoma; 17 patients had smoking histories. There were 32 patients in stage IVB and 4 patients in stage IIIB.
Table 1
| Characteristics | Parameter value | Percentage value, % |
|---|---|---|
| Age, years | Median: 62 | Range: 46–74 |
| Gender | ||
| Male | 18 | 50.0 |
| Female | 18 | 50.0 |
| Smoking history | ||
| Yes | 17 | 47.2 |
| No | 19 | 52.8 |
| Pathological diagnosis | ||
| Adenocarcinoma | 28 | 77.8 |
| Squamous cell carcinoma | 8 | 22.2 |
| Pathological stage | ||
| IIIB | 4 | 11.1 |
| IVB | 32 | 88.9 |
| Total | 36 | 100.0 |
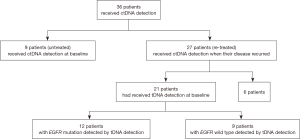
In the total 36 patients, 9 patients were untreated and received ctDNA detection before their primary treatment, and other 27 patients were re-treated and received ctDNA detection when their disease began to progress in therapeutic process. Out of the 27 re-treated patients, 21 patients had ever received tissue DNA (tDNA) detection at baseline. The schematic of enrolled patients is showed in Figure 1.
Actionable aberrations
Following with the deep hybrid-captured sequencing, we filtered the patients’ variants with a gene database which was established by 1Gene Bio-tech Co., Ltd. and contained 66 drug-associated genes in lung cancer. Corresponding results are exhibited in Table 2, totally 43 actionable aberrations were discovered in 27 patients (75%, 27/36, 22 were re-treated and 5 were untreated), including 21 cases of adenocarcinoma and 6 cases of squamous cell carcinoma. Gene EGFR had the highest mutational frequency with 36.1% (13/36), and the subsequent mutational genes were PTEN (13.9%, 5/36), MAP2K1 (11.1%, 4/36), KRAS (11.1%, 4/36), PIK3CA (8.3%, 3/36), GNAQ (5.6%, 2/36) and BRAF (5.6%, 2/36). When considering pathological type, EGFR was also the most frequent mutational gene in the 28 patients with adenocarcinoma (42.86%, 12/28), followed with KRAS (17.86%, 5/28), MAP2K1 (10.71%, 3/28), PTEN (10.71%, 3/28), PIK3CA (7.14%, 2/28), BRAF (7.14%, 2/28), HRAS (3.57%, 1/28). In the 8 patients with squamous cell carcinoma, the most frequent mutations were PTEN and GNAQ, with the same rate 25% (2/8), and EGFR, PIK3CA, KIT also had the same variant frequency of 12.5% (1/8). None of patients was detected with the mutated fusion in gene ALK or ROS1 in this study.
Table 2
| Patient ID | Gene | c.anno | p.anno | Pathological type | Treatment status | Subsequent treatment |
|---|---|---|---|---|---|---|
| 10267 | EGFR | c.2573T>G | p.L858R | AC | RE | Palliative care |
| 10395 | EGFR | c.2236_2250del15 | p.E746_A750delELREA | AC | RE | TKI |
| 10422 | PTEN | c.697C>T | p.R233* | AC | RE | Chemotherapy |
| 10430 | KRAS | c.38G>A | p.G13D | AC | RE | Palliative care |
| 10506 | EGFR | c.2369C>T | p.T790M | AC | RE | TKI |
| 10507 | MAP2K1 | c.167A>C | p.Q56P | AC | RE | TKI |
| 10512 | MAP2K1 | c.157T>C | p.F53L | AC | RE | TKI |
| 10551 | GNAQ | c.548G>A | p.R183Q | SC | RE | Palliative care |
| 10783 | PIK3CA | c.3140A>T | p.H1047L | SC | RE | TKI |
| 10789 | KRAS | c.34G>A | p.G12S | AC | RE | Palliative care |
| 10823 | EGFR | c.2236_2250del15 | p.E746_A750delELREA | AC | RE | TKI |
| 10864 | PIK3CA | c.1624G>A | p.E542K | AC | RE | Palliative care |
| 10877 | EGFR | c.2573T>G | p.L858R | AC | RE | TKI |
| 11085 | EGFR | c.2240_2254del15 | p.L747_T751delLREAT | AC | RE | TKI |
| 10836 | EGFR | c.2236_2250del15 | p.E746_A750delELREA | AC | UN | † |
| 10040 | PTEN | c.389G>A | p.R130Q | SC | UN | † |
| 10184 | KRAS | c.182A>T | p.Q61L | AC | RE | Chemotherapy |
| PTEN | c.388C>T | p.R130* | ||||
| 10257 | EGFR | c.2573T>G | p.L858R | AC | RE | TKI |
| EGFR | c.2369C>T | p.T790M | ||||
| 10312 | BRAF | c.1799T>A | p.V600E | AC | RE | Palliative care |
| PTEN | c.388C>T | p.R130* | ||||
| 10792 | EGFR | c.2582T>A | p.L861Q | SC | RE | TKI |
| EGFR | c.2369C>T | p.T790M | ||||
| 10843 | EGFR | c.2369C>T | p.T790M | AC | RE | TKI |
| EGFR | c.2240_2257del18 | p.L747_P753delinsS | ||||
| 10415 | EGFR | c.2573T>G | p.L858R | AC | RE | TKI |
| EGFR | c.2369C>T | p.T790M | ||||
| 11088 | KRAS | c.35G>T | p.G12V | AC | RE | TKI |
| MAP2K1 | c.199G>A | p.D67N | ||||
| 11095 | GNAQ | c.548G>A | p.R183Q | SC | RE | Palliative care |
| PTEN | c.697C>T | p.R233* | ||||
| 11353 | EGFR | c.2155G>A | p.G719S | AC | UN | † |
| EGFR | c.2582T>A | p.L861Q | ||||
| KRAS | c.37G>A | p.G13S | ||||
| 10064 | BRCA1 | c.3548A>G | p.K1183R | SC | UN | † |
| BRCA1 | c.2612C>T | p.P871L | ||||
| KIT | c.1676T>G | p.V559G | ||||
| MAP2K1 | c.332T>G | p. I111S | ||||
| 10558 | BRAF | c.1397G>T | p.G466V | AC | UN | † |
| EGFR | c.2235_2249del15 | p.E746_A750delELREA | ||||
| HRAS | c.182A>G | p.Q61R | ||||
| PIK3CA | c.1624G>A | p.E542K | ||||
| 10273 | – | – | – | AC | UN | † |
| 10423 | – | – | – | AC | UN | † |
| 11103 | – | – | – | SC | UN | † |
| 10248 | – | – | – | AC | UN | † |
| 10271 | – | – | – | AC | RE | TKI |
| 11106 | – | – | – | SC | RE | Palliative care |
| 11097 | – | – | – | AC | RE | Chemotherapy |
| 10921 | – | – | – | AC | RE | TKI |
| 10503 | – | – | – | AC | RE | TKI |
AC, adenocarcinoma; SC, squamous cell carcinoma; RE, re-treated; UN, untreated; c.anno, cDNA annotation; p.anno, protein annotation; *, nonsense mutation; –, negative; †, the treatment of newly diagnosed patients were not included.
The phenomenon of multiple mutations was discovered in 11 patients, including 8 patients with double gene mutations, 1 patient with three gene mutations, and 2 patients with four gene mutations. Among these patients, we observed that aberrations usually appeared with an accompaniment of EGFR mutation (55%, 6/11), and we speculated that EGFR mutation may be one of the primary founding clusters of lung cancer. Twenty-two out of 27 re-treated patients (81.5%, 22/27) were identified with pathogenic mutations, and all the mutational genes are located in the protein kinase signaling pathways. Furthermore, 5 patients were characterised with the EGFR p.T790M aberration which is known as the main cause of tyrosine kinase inhibitor TKI) drug resistance in NSCLC. In summary, ctDNA detection technology provides us with a novel helpful method to study the pathogenic mechanisms, drug resistance, and disease recurrence in NSCLC.
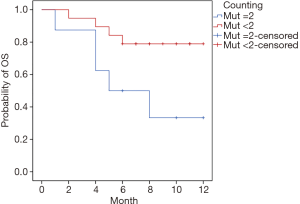
It was on the time when disease began to progress that the 27 re-treated patients received ctDNA detection. Results pointed out that 8 patients had double-gene mutations (mutation count =2) and 19 patients had non-gene mutation or single-gene mutation (mutation count <2) (Table 2). Subsequent therapeutic strategy was partially based on the genomic information. The survival follow-up was continuing to April 30, 2018, and it showed that a total of 9 patients were dead, including 5 patients in group “mutation count =2” and 4 patients in group “mutation count <2”. The 12-month survival rate of group “mutation count =2” and group “mutation count <2” was 50.00% (4/8) and 84.21% (16/19) respectively, and there was a statistically significant difference between the two groups (7.08±1.43 vs. 10.37±0.74 months, P=0.034; Figure 2).
Correlation between EGFR status and clinicopathologic characteristics
The correlation between mutational status of EGFR and clinicopathologic features was analyzed. Results showed that EGFR mutations usually occurred in younger patients in this study, the positive mutational rate of patients aged under 60 and above 60 was 57.1% and 22.7% respectively (P<0.05). There were no statistical differences between EGFR status and other clinical features, such as gender, pathological diagnosis, smoking history, pathological stage, and primary treatment (Table 3).
Table 3
| Variables | EGFR-mut | EGFR-wt | P |
|---|---|---|---|
| Age, years | 0.036 | ||
| <60 | 8 | 6 | |
| ≥60 | 5 | 17 | |
| Gender | 0.298 | ||
| Female | 8 | 10 | |
| Male | 5 | 13 | |
| Pathological diagnosis | 0.093 | ||
| AC | 12 | 16 | |
| SC | 1 | 7 | |
| Smoking history | 0.137 | ||
| Ever | 4 | 13 | |
| Never | 9 | 10 | |
| Pathological stage | 1.000 | ||
| IIIB | 1 | 3 | |
| IVB | 12 | 20 | |
| Primary treatment | 1.000 | ||
| Yes | 3 | 6 | |
| No | 10 | 17 |
AC, adenocarcinoma; SC, squamous cell carcinoma; mut, mutation; wt, wild type.
In the process of analysis, we also found that all the squamous cell carcinoma patients (100%, 8/8) and 47.4% (9/19) adenocarcinoma patients had smoking history. There was a significant relation between pathological diagnosis and smoking history (P<0.001), indicating that people who have smoking habit are more likely to suffer squamous cell carcinoma than adenocarcinoma.
EGFR targeted therapy
We also compared the differences of mutational characteristics between ctDNA detection and tDNA detection among the 21 patients who had received a tDNA detection at baseline. It was found that the mutational characteristics had changed in the majority of patients (85.7%, 18/21) from baseline to disease progression (Table 4). In the 9 patients who had been classified as EGFR-wt (wild type) by tDNA detection, 8 patients (88.9%, 8/9) were detected out of genomic mutations by subsequent ctDNA detection, including 3 with EGFR mutations (33.3%, 3/9), 3 respectively with KRAS, PIK3CA, MAP2K1 mutation (11.1%, 1/9), 1 with BRAF, PTEN double-gene mutations (11.1%, 1/9), 1 with KRAS, PTEN double-gene mutations (11.1%, 1/9). In the 12 patients who were primary EGFR-mut (mutation) confirmed by tDNA detection, 3 were detected with non-mutations (25%, 3/12), 4 were found accompanying with a new mutational loci p.T790M in EGFR (33.3%, 4/12), 3 had different mutational genes (25%, 3/12), 2 were the same as primary (16.7%, 2/12), according to the ctDNA detection (Table 4).
Table 4
| Patient ID | Mutational status | Using targeted drugs | Using chemotherapeutic drugs | Time to progression (month) | |
|---|---|---|---|---|---|
| At baseline | At disease progression | ||||
| 10257 | EGFR L858R | EGFR L858R; EGFR T790M | Y | Y | 8 |
| 10843 | EGFR Ex19 Del | EGFR Ex19 Del; EGFR T790M | Y | Y | 3 |
| 10415 | EGFR L858R | EGFR L858R; EGFR T790M | Y | Y | 31 |
| 10506 | EGFR Ex19 Del | EGFR T790M | Y | N | 14 |
| 11088 | EGFR Ex19 Del | KRAS G12V; MAP2K1 D67N | Y | Y | 53 |
| 10512 | EGFR Ex19 Del | MAP2K1 F53L | Y | N | 7 |
| 10422 | EGFR L858R | PTEN R233* | Y | N | 38 |
| 10823 | EGFR Ex19 Del | EGFR Ex19 Del | Y | N | 7 |
| 10877 | EGFR Ex19 Del; EGFR L858R | EGFR L858R | Y | Y | 14 |
| 10503 | EGFR Ex19 Del | – | Y | N | 16 |
| 10271 | EGFR Ex19 Del | – | Y | Y | 5 |
| 10921 | EGFR G719X | – | Y | Y | 20 |
| 11106 | – | – | N | Y | 8 |
| 10864 | – | PIK3CA E542K | N | Y | 16 |
| 10267 | – | EGFR L858R | N | Y | 5 |
| 10430 | – | KRAS G13D | N | Y | 2 |
| 10312 | – | BRAF V600E; PTEN R130* | N | Y | 12 |
| 10184 | – | KRAS Q61L; PTEN R130* | N | Y | 11 |
| 10395 | – | EGFR Ex19 DEL | N | Y | 3 |
| 11085 | – | EGFR Ex19 DEL | N | Y | 4 |
| 10507 | – | MAP2K1 Q56P | N | Y | 5 |
*, nonsense mutation; –, negative; Ex, exon; Del, deletion; N, no; Y, yes.
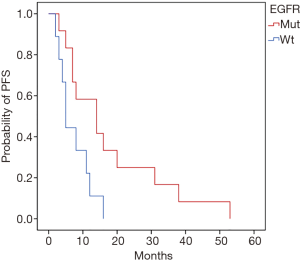
Based on the tDNA detection, we classified the 21 patients into two groups according to the status of EGFR. The subsequent therapeutic strategies were adopted according to the classification, 12 patients in group EGFR-mut were subsequently treated with EGFR-TKI or EGFR-TKI combined chemotherapy (some patients gave up TKI halfway because of poor tolerance or costly expense and adopted chemotherapy alternatively), and other 9 patients in group EGFR-wt were treated with platinum chemotherapeutic drugs (Table 4). The comparative analysis of PFS (progression free survival) between the two groups in the next 5 years revealed that patients with EGFR-mut had a significantly longer stable stage than those with EGFR-wt (18.00±4.41 vs. 7.33±1.58 months, P=0.024) (Figure 3).
DiscussionOther Section
NSCLC is the most common type and accounting for 80% of lung cancer, and the two main pathological classifications of NSCLC are adenocarcinoma and squamous cell carcinoma (15). Tissue biopsy is the gold standard for the diagnosis of lung cancer in the current mainstream views. However, with the development of liquid biopsy technology, ctDNA detection, which was used as a complementary test for tDNA detection, has become more and more clinically significant for patients whose tissue samples were unobtainable (16). CtDNA is a kind of cell-free DNA which comes from the genome of tumor cells and circulating in peripheral blood. A couple of studies have proven that ctDNA is of great concordance with tumor tissue DNA in sensitivity and specificity of tumor associated DNA mutations (17-19).
In this study, deep targeted sequencing was performed to reveal the gene mutations of 36 advanced NSCLC patients by their ctDNA samples. Results indicated that 27 patients had at least one mutational gene, and the most common mutational gene was EGFR, with a frequency of 36.11%, including 42.86% in patients with adenocarcinoma and 12.5% with squamous cell carcinoma. Our finding is in consistent with previous studies which reported that the mutational frequency of EGFR in NSCLC detected in ctDNA ranges from 20.4% to 43.0% (20,21).
The total mutational frequency of aberrations in EGFR was the highest among all the therapeutic genetic targets in Oriental population. Previous studies found that the mutational frequency of EGFR in lung adenocarcinoma range from 47.9% to 55.7%, while it is much lower in lung squamous cell carcinoma (22). Similarly, our result also showed that the mutational frequency of EGFR in adenocarcinoma and squamous cell carcinoma was 42.86% and 12.5% respectively. In this study, EGFR mutations usually occurred in younger patients with age ≤60 (P<0.05), and there was no statistical significance in EGFR status and other clinical features, which was different from other studies probably because of the limited sample size (23,24). At the same time, our analysis proved that smoking had significant statistical correlation with pathological type (P<0.001) in advanced NSCLC, indicating that smokers are more likely to develop into squamous cell carcinoma but not adenocarcinoma.
Multiple mutations were rarely reported in previous clinical studies of lung cancer, and the majority of reported mutational frequency ranged 3.13% to 8.33% (25,26). There were 11 out of 36 patients in this study had been detected with multiple mutations, with a frequency of 30.56%, which was higher than previous studies. It was speculated that the enrolled 36 patients were all advanced cases and most of them underwent disease progression and drug resistance after primary treatment, which might lead to the appearance of new mutations. On the other hand, limited sample size and the biased composition of pathological type might also lead to the different outcomes.
The results of ctDNA detection which performed to the 27 re-treated patients showed that 8 patients were classified as double-gene mutations and 19 patients were single-gene mutation or negative mutation. Subsequently clinical medication was also determined partially according to the genomic status, and the overall survival of patients with double-gene mutations was significant shorter than patients with single-gene mutation or non-gene mutation (P=0.034), suggesting that ctDNA may serve as one of the indicators of prognosis in the future.
The survival analysis of the 21 patients who were detected with EGFR mutations by tissue biopsy revealed that EGFR-mut patients treated with EGFR-TKI or EGFR-TKI combined chemotherapy had a significantly longer progression free survival than EGFR-wt patients treated with chemotherapeutics only (P=0.024). Therefore, the detection of EGFR mutation in NSCLC patients showed important clinical significance during treatment sessions (27-29). Subsequently, a ctDNA detection was conducted to the above 21 patients when they showed a disease progression or drug resistance. Results revealed that the mutational spectrum had changed in the majority of them (85.7%, 18/21), and it’s speculated that the changes of mutational spectrum might lead to those bad outcomes, though more validation tests need to be performed. All these results suggest us that it is useful to re-perform gene detection for the following disease monitoring and drug selection for patients who were undergoing disease progression or drug resistance.
ConclusionsOther Section
In summary, this study revealed the molecular mutational characteristics of advanced NSCLC patients with post-treatment disease progression by next-generation sequencing based ctDNA detection, and proved the capability of molecular diagnosis by liquid biopsy for advanced lung cancer patients. When comparing the results between baseline tDNA detection and post-treatment ctDNA detection, we found that the genomic mutational spectrum had changed in the majority of the patients who underwent disease progression or drug resistance. Patients with multiple mutations obtained a shorter overall survival than those with less mutations, which indicates that multiple genes mutation may indicate a poor prognosis. Therefore, it is necessary to take a gene detection for therapeutic options and disease progression monitoring for NSCLC patients.
AcknowledgmentsOther Section
Funding: None.
FootnoteOther Section
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.20). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Zhejiang Cancer Hospital (IRB-2018-201), and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Preusser M, Capper D, Ilhan-Mutlu A, et al. Brain metastases: pathobiology and emerging targeted therapies. Acta Neuropathol 2012;123:205-22. [Crossref] [PubMed]
- Ostrom QT, Wright CH, Barnholtz-Sloan JS. Brain metastases: epidemiology. Handb Clin Neurol 2018;149:27-42. [Crossref] [PubMed]
- Gállego Pérez-Larraya J, Hildebrand J. Brain metastases. Handb Clin Neurol 2014;121:1143-57. [Crossref] [PubMed]
- Vicent S, Perurena N, Govindan R, et al. Bone metastases in lung cancer. Potential novel approaches to therapy. Am J Respir Crit Care Med 2015;192:799-809. [Crossref] [PubMed]
- Sun Y, Guan Z, Liao M, et al. Expert consensus on the diagnosis and treatment of bone metastasis in lung cancer (2014 version). Zhongguo Fei Ai Za Zhi 2014;17:57-72. [PubMed]
- Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol 2013;31:1097-104. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139-42. [Crossref] [PubMed]
- Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol 2014;25:2304-13. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Zhao H, Chen KZ, Hui BG, et al. Role of circulating tumor DNA in the management of early-stage lung cancer. Thorac Cancer 2018;9:509-15. [Crossref] [PubMed]
- Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [Crossref] [PubMed]
- Ikeda S, Schwaederle M, Mohindra M, et al. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J Hematol Oncol 2018;11:76. [Crossref] [PubMed]
- Davidson MR, Gazdar AF, Clarke BE. The pivotal role of pathology in the management of lung cancer. J Thorac Dis 2013;5:S463-78. [PubMed]
- Hicks JK, Saller J, Wang E, et al. Cell-free circulating tumor DNA supplementing tissue biopsies for identification of targetable mutations: Implications for precision medicine and considerations for reconciling results. Lung Cancer 2017;111:135-8. [Crossref] [PubMed]
- Lebofsky R, Decraene C, Bernard V, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015;9:783-90. [Crossref] [PubMed]
- Xu S, Lou F, Wu Y, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett 2016;370:324-31. [Crossref] [PubMed]
- Ma M, Shi C, Qian J, et al. Comparison of plasma and tissue samples in epidermal growth factor receptor mutation by ARMS in advanced non-small cell lung cancer. Gene 2016;591:58-64. [Crossref] [PubMed]
- Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. [Crossref] [PubMed]
- Nie K, Jia Y, Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol 2015;36:7-19. [Crossref] [PubMed]
- Gou LY, Wu YL. Prevalence of driver mutations in non-small-cell lung cancers in the People's Republic of China. Lung Cancer (Auckl) 2014;5:1-9. [PubMed]
- Normanno N, Denis MG, Thress KS, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017;8:12501-16. [Crossref] [PubMed]
- Boch C, Kollmeier J, Roth A, et al. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC): routine screening data for central Europe from a cohort study. BMJ Open 2013;3: [Crossref] [PubMed]
- Li S, Li L, Zhu Y, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer 2014;110:2812-20. [Crossref] [PubMed]
- Wen YS, Cai L, Zhang XW, et al. Concurrent oncogene mutation profile in Chinese patients with stage Ib lung adenocarcinoma. Medicine (Baltimore) 2014;93:e296. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Kim ST, Sung JS, Jo UH, et al. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Med Oncol 2013;30:328. [Crossref] [PubMed]