
Alternatively activated NUSAP1 promotes tumor growth and indicates poor prognosis in hepatocellular carcinoma
Introduction
Primary liver cancer is the fifth most common cancer type around the world, with increasing incidence in the past several decades (1). According to the statistics from GLOBOCAN 2012, approximately 782,500 new cases and 745,500 deaths occurred globally due to liver cancer; however, approximately 50% of all cases were in China (2). Previous studies have found that the high incidence of primary liver cancer in China was associated with the increasing prevalence of chronic infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) (2-5). Hepatocellular carcinoma (HCC), originating in hepatocytes, is the major histologic subtype of liver cancer and accounts for ~80% of primary liver cancer cases. Currently, there are various types of HCC therapies, such as hepatic resection, chemotherapy, immunotherapy, radiofrequency ablation or alcohol ablation, and liver transplantation (6-8). Although these therapies have improved the survival of HCC patients compared to that observed in the last decade, the overall prognosis is still poor. Thus, underlying molecular mechanism investigation, novel biomarker identification and recurrence prediction are urgent goals to improve the early diagnosis and prognosis of HCC (7).
Microarrays can detect hundreds to millions of different molecules at the same time. Microarray analysis is a rapid technique that plays an indispensable role in the human genome project (HGP). With advancements in science and scientific techniques, microarray analysis has been employed for various studies, for example, large-scale genotyping, new gene discovery, mutation identification, and, in particular, cancer genome studies (9). Microarray analysis has become one of the major methods for finding the origin and progression of cancer using data from the whole genome (10). Currently, microarrays are successfully being used in the discovery of new biomarkers in most human cancers, such as breast cancer (11), prostate cancer (12), lung cancer (13) and HCC (14). Although gene profiling is a powerful tool, the differentially identified genes (DEGs) identified in one study often cannot be identified in another study and can easily be influenced by confounding factors either because of sample selection or due to different analysis platforms. Hence, genome-wide meta-analyses are necessary to summarize similar gene-based data from multiple databases.
In this study, we performed an integrated analysis based on HCC mRNA profiles from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases to identify potential biomarkers of HCC on a genome-wide scale. Eleven proliferation-related DEGs (BBOX1, CENPA, DLGAP5, FOXM1, KIF20A, KIF4A, MKI67, NCAPG, NUSAP1, SFN, and TPX2) were identified at the in silico stage. Among these DEGs, nucleolar and spindle-associated protein 1 (NUSAP1) gene was one of the overexpressed genes in HCC and displayed a high likelihood of being involved in HCC progression. This study mainly focused on exploring the effects of NUSAP1 in HCC.
Methods
Microarray data collection, comparison and meta-analysis
Because small sample sizes and different microarray analyses may lead to errors during the screening process, the results may not be completely reliable. A meta-analysis is a statistical analysis method that combines multiple studies to analyze a larger population to obtain more accurate and comprehensive results. The GEO (https://www.ncbi.nlm.nih.gov/geo/) is a national public database that includes high-throughput gene expression, array- and sequence-based data (15). In this study, eligible gene expression data were obtained from the GEO (Affymetrix Human Genome U133 Plus 2.0 Array). Then, a meta-analysis was performed to identify the DEGs based on these datasets.
TCGA data extraction and analysis
After identifying DEGs using the method described above, we analyzed the effects of the genes in patients and their association with survival. mRNA expression and clinical data from TCGA were extracted from cBioPortal (www.cbioportal.org). Then, the data were preprocessed and analyzed by R. A violin plot was constructed to show the differences in specific gene expression between HCC tissues and nontumor tissues. Kaplan-Meier plots were constructed to demonstrate the relationship between the dysregulated genes and patient survival. In addition, various clinicopathological characteristics, including age at initial pathologic diagnosis, sex, TNM stage, histologic grade and pathologic stage, were investigated. The data was quantified and analyzed in R, and Chi-square test was performed.
Short hairpin RNA (shRNA) sequencing and data analysis
shRNA sequencing was conducted to investigate differential gene expression in HCC by knocking down genes with shRNAs. The shRNA sequencing data were uploaded and analyzed using a website (https://ncedu.shinyapps.io/shrna/). MDS, BCV, MAPlot, and two files (QC.csv and CPM.csv) were obtained from the website. Then, logFC and adjust P values [−log10(P)] were plotted in a volcano plot by using Excel 2016. To elucidate the possible molecular mechanisms regulated by the DEGs in HCC, we conducted gene set enrichment analysis (GSEA) to identify significantly enriched pathways. Data were uploaded and interpreted using the WEB-based GEne SeT AnaLysis Toolkit (http://www.webgestalt.org/option.php).
shRNA sequence construction and transfection
shNUSAP1 and shFoxM1 expression vectors were purchased from GeneChem (Shanghai, China). The target sequences of the above shRNA expression constructs are as follows: shNUSAP1#1: CCGGGAGGGCAACCAAGTTGTTAAACTCGAGTTTAACAACTTGGTTGCCCTCTTTTTTG; shNUSAP1#2: CCGGGAGCACCAAGAAGCTGAGAATCTCGAGATTCTCAGCTTCTTGGTGCTCTTTTTTG; and shNUSAP1#3: CCGGGAACCACACAAAGGAAAGCTACTCGAGTAGCTTTCCTTTGTGTGGTTCTTTTTTG. The positive control shFoxM1 sequence is as follows: CCGGGCCAATCGTTCTCTGACAGAACTCGAGTTCTGTCAGAGAACGATTGGCTTTTT.
Transient transfection of cells was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Cell proliferation assay
HCC cells were cultured in DMEM with 10% (v/v) FBS (Gibco). Culture flasks were placed in an incubator (37 °C, 5% CO2) and subcultured every 3 days. To perform cell proliferation assay, we divided HCC cells into 24-well plates at a density of 5×103 cells/well and cultured them with 0.5 mL DMEM (with 10% FBS) for 24 h. Then, the cells were transfected with constructed shRNA expression vectors. After transfection, the bromodeoxyuridine (BrdU) assay was performed according to the manufacturer’s protocol for the BrdU colorimetric cell proliferation assay kit (cat# 1647229; Roche). For clonogenic assay, HCC cells (3×103 per well) were plated in 6-well plates and cultured for 2 weeks. All plates were stained with 0.5% crystal violet (w/v), and the colony numbers were determined. Cell cycle distribution was analyzed by propidium iodide staining and flow cytometry.
RNA purification and quantitative RT-PCR
For real-time PCR, total RNA (1 µg) was subjected to reverse transcription using a reverse transcription reagent kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time PCR was performed with an ABI Step Plus One Sequence Detection System (Applied Biosystems). The specific primers used for PCR amplification are as follows: NUSAP1-F: AAGCAAGTTTGTCTCGTCC, NUSAP1-R: GAGATGGGGTTGTTTGTAAG, GAPDH-F: TGACTTCAACAGCGACACCA, and GAPDH-R: CACCCTGTTGCTGTAGCCAAA. All experiments were repeated at least three times with consistent results.
Statistical analysis
Data were assessed using Student’s t-test or one-way ANOVA followed by Fisher’s multiple range test.
Results
Integrated analysis identified 11 proliferation-related genes that were significantly overexpressed in HCC
In this study, we identified five publicly available HCC datasets (GSE14520, GSE41804, GSE45267, GSE60502 and GSE6764) and manually extracted the data for 650 liver samples (including 341 HCC and 309 normal tissues; Table S1) from the GEO database. To scan for robust HCC-associated biomarkers across multiple independent cohorts, we performed two meta-analyses to detect DEGs between HCC and normal tissues. In total, 580 candidates represented by 13,021 transcripts displayed a considerable change in expression (P<0.0001) in at least one meta-analysis. As shown in Figure 1A, many DEGs (6.38%, 37/580) were common across both analyses. This number was over seven times higher than that expected from each analysis (5 genes, χ2 test, P<10−4). This result indicated the reliability of the meta-analyses in the detection of biomarkers of HCC.
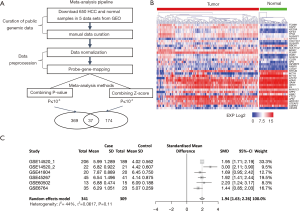
An additional independent dataset from TCGA provided an opportunity for confirming the primary biomarkers related to HCC. Only one gene (CLGN) did not show significant change in expression (P>0.0001) between 373 HCC and 50 normal tissues and was thus removed from the further analysis. Interestingly, among the 36 common DEGs, most (11 genes) were involved in “cell cycle and division”. Most importantly, all of these proliferation-related DEGs were upregulated in HCC tissues (Figure 1B), and most of them had been demonstrated to have crucial roles in HCC progression [including FOXM1 (16), CAP2 (17), KIF20A (18), KIF4A (19) and DLGAP5 (20)] in previous reports. Therefore, based on an integrated analysis of 6 independent HCC datasets (5 GEO datasets and 1 TCGA dataset) consisting of 714 HCC cases and 359 normal liver samples, we identified eleven proliferation-related DEGs (BBOX1, CENPA, DLGAP5, FOXM1, KIF20A, KIF4A, MKI67, NCAPG, NUSAP1, SFN, and TPX2) that were significantly and consistently overexpressed in HCC tissues across all datasets. Although we speculated that NUSAP1 likely participates in HCC development and progression, the function of NUSAP1 in HCC remained unknown.
Overexpression of NUSAP1 in HCC is related to clinicopathological features
NUSAP1, a 55-kDa microtubule-associated protein (MAP), is one of the members essential for the arrangement and functioning of the mitotic spindle (21). Our meta-analysis based on the five GEO datasets indicated that the expression level of NUSAP1 in HCC was higher than that in the nontumor tissues (P=1.87×10−73). In addition, forest plots illustrated that these results were stable without heterogeneity (I2=44%, τ2=0.0617, P=0.11, Figure 1C). Furthermore, TCGA transcriptome data confirmed the significant differential expression of NUSAP1 between tumor and nontumor tissues (t-test =8.93, P=7.17×10−16, Figure 2A).
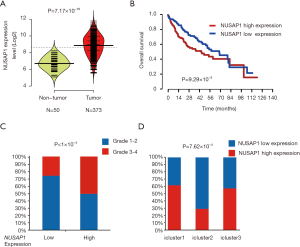
To explore the relationship between NUSAP1 and HCC clinicopathological features, we analyzed the expression profiles of HCC patients and clinical data from TCGA. In total, 336 patients were included in this analysis, and the cutoff value of NUSAP1 expression was defined by the median expression. As shown in Figure 2B, high expression of NUSAP1 was significantly associated with poor survival in HCC patients (P=0.00929). In addition, we explored the relationships between the expression level of NUSAP1 and clinicopathological features in HCC (Table 1). The data implied that high NUSAP1 expression was associated with tumor histologic grade (P<1×10−4, Figure 2C), tumor TNM stage (P=0.044), and patient age (P=0.018, Table 1).
Table 1
| Clinicopathologic measurement data | NUSAP1 expression (n=336) | |||||
|---|---|---|---|---|---|---|
| Low (n=168) | High (n=168) | P value | ||||
| Count | % | Count | % | |||
| Age | 0.018 | |||||
| ≤50 years | 28 | 8.3 | 46 | 13.7 | ||
| >50 years | 140 | 41.7 | 122 | 36.3 | ||
| Gender | 0.060 | |||||
| Male | 123 | 36.6 | 107 | 31.8 | ||
| Female | 45 | 13.4 | 61 | 18.2 | ||
| TNM stage | 0.044 | |||||
| Stage I–II | 134 | 39.9 | 118 | 35.1 | ||
| Stage III–IV | 34 | 10.1 | 50 | 14.9 | ||
| Histologic grade | 0.000 | |||||
| G1–G2 | 124 | 36.9 | 83 | 24.7 | ||
| G3–G4 | 44 | 13.1 | 85 | 25.3 | ||
TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; NUSAP1, nucleolar and spindle-associated protein 1.
Furthermore, NUSAP1 expression was associated with multiplatform integrative molecular subtyping performed by TCGA Research Network. Patients with different pathological and molecular characteristics were divided into three major groups (iCluster1, iCluster2 and iCluster3), and iCluster1 was directly correlated with the abnormal activation of cell proliferation (22). We integrated the expression profiles and iCluster data for HCC patients and found that patients with high NUSAP1 expression were enriched in iCluster1 (Figure 2D). This result implied that the high expression of NUSAP1 can activate cell proliferation.
In summary, high NUSAP1 expression plays a vital role in HCC occurrence and development and is closely associated with patient prognosis. NUSAP1 might thus be used as a cancer therapeutic target site in the future.
Downregulation of NUSAP1 suppresses the expression of genes involved in cell proliferation
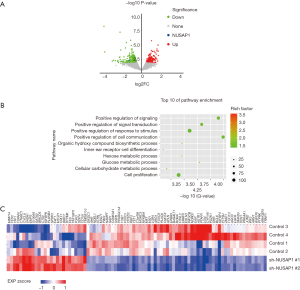
To investigate the biological significance of NUSAP1 expression in cancer cells, we downloaded RNA-Seq data from ENCODE to identify DEGs between HepG2_Control and HepG2_shNUSAP1 cells. Overall, 656 candidate genes, consisting of 264 upregulated genes and 392 downregulated genes, exhibited considerable change (P<0.05, Figure 3A). To further explore the biological significance of NUSAP1 in gene expression in HCC, we analyzed DEGs by overrepresentation analysis (ORA). As shown in Figure 3B, the DEGs were significantly enriched in 10 functional pathways (FDR <0.001), including 1 pathway that was related to cell proliferation and 3 pathways that were associated with metabolism. Most importantly, cell proliferation (GO:0008283) was included the largest number of candidate DEGs (85 genes, Figure 3C). Consistent with the clinicopathological data analysis, these results further confirmed that NUSAP1 plays vital roles in cell proliferation and cell cycle.
NUSAP1 promotes proliferation and cell cycle progression in HCC cells
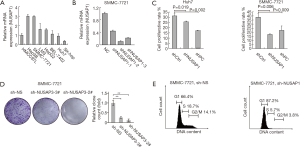
To explore the biological effects of NUSAP1 in HCC cell proliferation, we first detected NUSAP1 mRNA expression in 8 different HCC cell lines (HMCC97L, SMMC-7721, HepG2, Hep3B, BEL-7402, Huh7, SK-Hep1 and LM3). High expression of NUSAP1 was observed in SMMC-7721 and Hep3B cell lines, while low expression of NUSAP1 was detected in Huh7 and SK-Hep1 cell lines (Figure 4A). Subsequently, quantitative RT-PCR was used to evaluate the effectiveness of silencing NUSAP1 using three established shRNAs. shNUSAP1#2 could decrease the expression level of NUSAP1 to 11% (Figure 4B). SMMC-7721 and Huh7 cells were transfected with shNC, shNUSAP1 and shFoxM1 to establish shCtrl, shNUSAP1 and shPC groups, respectively. SMMC-7721 and Huh7 cells in the shCtrl group could divide normally (Figure 4C). FoxM1 is an indispensable gene for cell proliferation. Silencing of the FoxM1 gene in proliferating cells inhibited cell (Figure 4C). Knockdown of NUSAP1 suppressed cell proliferation in the two HCC cell lines (Figure 4C). Because the inhibition of proliferation in SMMC-7721 cells was the most obvious after NUSAP1 gene silencing, we further verified the results in colony forming and cell cycle analyses. As shown in Figure 4D,E, the growth of SMMC-7721 cells was significantly inhibited by NUSAP1 knockdown. Consistent with this result, NUSAP1 knockdown inhibited cell cycle progression and arrested most cells at the G1 phase. Therefore, NUSAP1 silencing can inhibit HCC cell growth and proliferation by blocking cell cycle progression.
Discussion
NUSAP1 is a recently identified 55-kDa MAP (23). Microtubules participate in the assembly of the mitotic spindle, which is crucial for ensuring the precise distribution of cellular material between two daughter cells (21). NUSAP1 is one of the proteins essential for mitotic spindle arrangement and function (24). By comparing gene expression patterns between proliferating and differentiating MC3T3E1 mouse osteoblasts in vitro, Raemaekers et al. [2003] identified NUSAP1 as a proliferation marker in cells (24). NUSAP1 expression is upregulated in dividing cells and reaches peak levels at G2 to M phase transition during the cell cycle (21,23,24). NUSAP1 overexpression leads to extended bundling of cytoplasmic microtubules during interphase. In contrast, the depletion of NUSAP1 in proliferating cells by RNA interference causes nuclear defects and disruptions in normal cell cycle progression (24). These data demonstrate the essential roles of NUSAP1 during cell proliferation.
Previous studies have found that NUSAP1 is actively expressed in some cancers such as prostate cancer (25), breast cancer (26), melanoma (27,28), and glioma (29), and NUSAP1 gene expression is associated with cell proliferation, migration and invasion in some specific cancer types (26,30). Gordon et al. [2017] found that NUSAP1 accelerates the invasion and metastasis of prostate cancer by partly modulating downstream FAM101B, which is a TGFβ1 signaling effector, and participates in epithelial-mesenchymal transition (EMT) (31,32). Additionally, researchers have found that NUSAP1 is regulated by the Rb-E2F pathway in prostate cancer (25). These findings all imply the vital functions of NUSAP1 in the development and progression of various tumors. Nevertheless, the mechanisms of NUSAP1 in HCC remains unknown.
In our study, meta-analysis of five independent datasets validated high mRNA expression of NUSAP1 in HCC in a large sample (650 samples). As shown in the forest plot (Figure 1C), compared to that in nontumor samples, NUSAP1 was predominantly overexpressed in HCC samples [SMD =1.94, 95% CI, 1.63–2.26, I2=44%]. Additionally, TCGA dataset confirmed the high expression of NUSAP1 in HCC (Figure 1B). On the other hand, the clinical data obtained from TCGA suggested that the overexpression of NUSAP1 in HCC is related to clinicopathological features (such as poor survival and high tumor histologic grade and TNM stage). Overexpression of NUSAP1 was also associated with the iCluster1 subgroup defined by TCGA Research Network, implying the NUSAP1 could activate cell proliferation.
In addition, we established a cell model with NUSAP1 downregulation to explore the role of NUSAP1 in HCC. Pathway analysis based on RNAseq data suggested that NUSAP1 could active the expression of genes involves in cell proliferation. As shown in Figure 4C, NUSAP1 expression was downregulated after the transfection of NUSAP1-targeting shRNA into two cell lines, and compared to that in the shCtrl-transfected cells, proliferation was dramatically inhibited in shNUSAP1-trasfected Huh7 and SMMC-7721 cells, indicating that NUSAP1 promotes cell proliferation in HCC. Based on the results of this study and previous reports, more experiments should be performed to investigate the role of NUSAP1 in the regulation of HCC development and progression, and NUSAP1 might be a target for HCC therapy.
In conclusion, compared to that in nontumor tissues, NUSAP1 is highly expressed in HCC tumor tissues. Analysis of clinical data showed that HCC patients with high NUSAP1 expression have poor prognosis. In addition, the results of cell proliferation assays also proved that NUSAP1 plays a crucial role in HCC cell proliferation. Our findings indicate that NUSAP1 might be a novel diagnostic marker and a therapeutic target in HCC.
Table S1
| GEO ID | Platforms | Total | Tumor | Non-tumor |
|---|---|---|---|---|
| GSE45267 | HG-U133_Plus_2 | 86 | 45 | 41 |
| GSE41804 | HG-U133_Plus_2 | 40 | 20 | 20 |
| GSE14520-2 | HG-U133A_2 | 43 | 22 | 21 |
| GSE14520-1 | HT_HG-U133A | 395 | 206 | 189 |
| GSE60502 | HG-U133A | 28 | 13 | 15 |
| GSE6764 | HG-U133_Plus_2 | 58 | 35 | 23 |
| Total | 650 | 341 | 309 |
HCC, hepatocellular carcinoma; GEO, Gene Expression Omnibus.
Acknowledgments
Funding: This work was supported in part by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Mas VR, Maluf DG, Archer KJ, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med 2009;15:85-94. [Crossref] [PubMed]
- McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis 2011;15:223-43. vii-x. [Crossref] [PubMed]
- Wang M, Wang Y, Feng X, et al. Contribution of hepatitis B virus and hepatitis C virus to liver cancer in China north areas: Experience of the Chinese National Cancer Center. Int J Infect Dis 2017;65:15-21. [Crossref] [PubMed]
- Greten TF, Sangro B. Targets for immunotherapy of liver cancer. J Hepatol 2017; [Epub ahead of print]. [PubMed]
- Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology 2002;122:1609-19. [Crossref] [PubMed]
- Duffy A, Greten T. Developing better treatments in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol 2010;4:551-60. [Crossref] [PubMed]
- Chaudhuri JD. Genes arrayed out for you: the amazing world of microarrays. Med Sci Monit 2005;11:RA52-62. [PubMed]
- Wadlow R, Ramaswamy S. DNA microarrays in clinical cancer research. Curr Mol Med 2005;5:111-20. [Crossref] [PubMed]
- Zheng T, Wang A, Hu D, et al. Molecular mechanisms of breast cancer metastasis by gene expression profile analysis. Mol Med Rep 2017;16:4671-7. [Crossref] [PubMed]
- Özdemir TR, Simsir A, Onay H, et al. Whole-genome gene expression analysis in urine samples of patients with prostate cancer and benign prostate hyperplasia. Urol Oncol 2017;35:607.e15-24. [Crossref] [PubMed]
- Zhang Y, Sui J, Shen X, et al. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of lung cancer. Oncol Rep 2017;37:3543-53. [Crossref] [PubMed]
- Lau WY, Lai PB, Leung MF, et al. Differential gene expression of hepatocellular carcinoma using cDNA microarray analysis. Oncol Res 2000;12:59-69. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res 2017;77:3135-9. [Crossref] [PubMed]
- Shibata R, Mori T, Du W, et al. Overexpression of cyclase-associated protein 2 in multistage hepatocarcinogenesis. Clin Cancer Res 2006;12:5363-8. [Crossref] [PubMed]
- Shi C, Huang D, Lu N, et al. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget 2016;7:26206-19. [PubMed]
- Hou G, Dong C, Dong Z, et al. Upregulate KIF4A Enhances Proliferation, Invasion of Hepatocellular Carcinoma and Indicates poor prognosis Across Human Cancer Types. Sci Rep 2017;7:4148. [Crossref] [PubMed]
- Liao W, Liu W, Yuan Q, et al. Silencing of DLGAP5 by siRNA significantly inhibits the proliferation and invasion of hepatocellular carcinoma cells. PLoS One 2013;8:e80789. [Crossref] [PubMed]
- Ribbeck K, Raemaekers T, Carmeliet G, et al. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr Biol 2007;17:230-6. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Iyer J, Moghe S, Furukawa M, et al. What's Nu(SAP) in mitosis and cancer? Cell Signal 2011;23:991-8. [Crossref] [PubMed]
- Raemaekers T, Ribbeck K, Beaudouin J, et al. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol 2003;162:1017-29. [Crossref] [PubMed]
- Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene 2013;32:70-7. [Crossref] [PubMed]
- Kretschmer C, Sterner-Kock A, Siedentopf F, et al. Identification of early molecular markers for breast cancer. Mol Cancer 2011;10:15. [Crossref] [PubMed]
- Ryu B, Kim DS, Deluca AM, et al. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One 2007;2:e594. [Crossref] [PubMed]
- Bogunovic D, O'Neill DW, Belitskaya-Levy I, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A 2009;106:20429-34. [Crossref] [PubMed]
- Marie SK, Okamoto OK, Uno M, et al. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer 2008;122:807-15. [Crossref] [PubMed]
- Bell R, Barraclough R, Vasieva O. Gene Expression Meta-Analysis of Potential Metastatic Breast Cancer Markers. Curr Mol Med 2017;17:200-10. [Crossref] [PubMed]
- Gordon CA, Gong X, Ganesh D, et al. NUSAP1 promotes invasion and metastasis of prostate cancer. Oncotarget 2017;8:29935-50. [Crossref] [PubMed]
- Gay O, Gilquin B, Nakamura F, et al. RefilinB (FAM101B) targets filamin A to organize perinuclear actin networks and regulates nuclear shape. Proc Natl Acad Sci U S A 2011;108:11464-9. [Crossref] [PubMed]


