
Effect of receptor for hyaluronan-mediated motility inhibition on radiosensitivity of lung adenocarcinoma A549 cells
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide. Approximately 70% of patients with lung cancer require radiotherapy to improve local control and survival. Despite its use to improve local control, radiotherapy has unsatisfactory overall curative effects because of tumor radioresistance (1) or low tolerance of the normal tissue to radiotherapy (2). Therefore, adjuvant radiosensitive treatments are needed to improve the therapeutic ratio, and novel biomarkers are needed to predict prognosis related to radiotherapy and radiosensitization.
Hyaluronic acid (HA) is an important extracellular matrix component that promotes cancer cell invasion through binding of cell surface receptors. Among the many factors that affect cancer cell invasion and metastasis, interactions between HA in the extracellular matrix and receptors on cancer cells have become an area of interest. Receptor for hyaluronan-mediated motility (RHAMM) is a soluble hyaluronan-binding protein that, with cluster of differentiation (CD)-44, serves as a major HA receptor. The RHAMM gene, which is located on human chromosome 5q33.2, contains 18 exons and encodes an 84-kDa protein (3). Alternative splicing of the RHAMM gene results in the generation of four different isoforms. This alternative splicing may be involved in the formation of hepatic carcinoma metastases (4). Based on location, RHAMM can be divided into intracellular and cellular surface subtypes: intracellular HA binding proteins and CD168, respectively. In human tissues, RHAMM can be detected by low mRNA expression in lung and pancreatic tissues, and high mRNA expression in the testes, thymus, and placenta (5). RHAMM expression is upregulated in several types of tumors, including stomach cancer (6), endometrial cancer (7), bladder cancer (8), prostate cancer (9), breast cancer (10), head and neck cancer (11), and glioblastoma (12).
Wang and colleagues (13) reported that RHAMM protein was expressed in 57% (89/156) of primary non-small cell lung cancer (NSCLC) cases, a number that correlated with those primary tumors that were locally advanced and poorly differentiated; it was also expressed in 96% (22/23) of cases of metastatic NSCLC. Survival analysis has shown that higher RHAMM mRNA expression predicts poor outcomes in patients with lung adenocarcinoma, but not in those with squamous cell carcinoma (13). Furthermore, RHAMM knockdown inhibits the migratory ability of human lung adenocarcinoma cell lines (H1975 and H3255). A recent study showed that RHAMM protein expression is a negative prognostic factor for large cell lung cancer (14). Thangavel et al. (15) reported that genetically modulating or pharmacologically inhibiting RHAMM activity could be used to treat metastatic phenotypes induced by retinoblastoma tumor suppressor protein loss in prostate cancer. Their findings suggest that RHAMM may be a treatment target in advanced prostate cancer.
Schütze and colleagues (16) have demonstrated that RHAMM mRNA expression in breast biopsy tissues is negatively correlated with tumor grade and overall survival. Furthermore, RHAMM splice variants associated with p53 mutations could be used to predict the susceptibility of breast cancer cells to radiotherapy; pharmacological inhibition of hyaluronan, the major ligand of RHAMM, increased the radiosensitivity of breast cancer cell lines.
RHAMM is a valuable prognostic factor in lung adenocarcinoma and may serve as a therapeutic target for the prevention of NSCLC metastasis (13). For select patients with breast cancer, response to radiotherapy may be improved by pharmaceutical strategies directed against RHAMM and its ligand HA (16). RHAMM is related to ERK and is crucial in ERK phosphorylation to its active form (17). The ERK1/2/MAPK pathway is a classical signaling system that mediates ligand-stimulated signals for the induction of cell proliferation, differentiation, and survival (18). In the present study, we hypothesized that RHAMM was a prognostic factor for radiotherapy effectiveness in patients with lung cancer and a target for radiosensitization. Lung adenocarcinoma cell line A549 was used to investigate the effect of inhibiting RHAMM gene expression on radiosensitivity and to determine its mechanisms of action related to ERK1/2 regulation.
Methods
NSCLC cell preparation
Human lung adenocarcinoma cell line A549 and large cell cancer cell line H460 were obtained from the Heilongjiang Provincial Institute of Oncology (Heilongjiang, China). Cells were cultured in 1,640 mediums (Hyclone, GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (Hangzhou Sijiqing Biological Engineering Materials Co., Ltd., Hangzhou, China) and cultured in an incubator at 37 °C with 5% CO2. The cells in the logarithmic growth phase were used for subsequent experiments.
RHAMM gene silencing
Small interfering (si) RNA sequences targeting all RHAMM splice variants (Suzhou Gemma Gene, Suzhou, China) including RHAMM-siRNA 1 (sense, 5'-AGG CUA AAU GCU GCA CUA ATT-3'; antisense, 5'-UUA GUG CAG CAU UUA GCC UTT-3'), RHAMM-siRNA 2 (sense, 5'-GCU AGA UAU UGC CCA GUU ATT-3'; antisense, 5'-UAA CUG GGC AAU AUC UAG CTT-3'), RHAMM-siRNA 3 (sense, 5'-GCA AAC ACU GGA UGA GCU UTT-3'; antisense, 5'-AAG CUC AUC CAG UGU UUG CTT-3'), and nonspecific siRNA (sense, 5'-UUC UCC GAA CGU GUC ACG UTT-3'; antisense, 5'-ACG UGA CAC GUU CGG AGA ATT-3') were used for cell transfection. One day prior to transfection, cells were cultured overnight in antimicrobial-free media to achieve the desired 30% to 50% confluency.
First, X-tremeGENE siRNA transfection reagent (Roche Diagnostics GmbH, Mannheim, Germany) and siRNA with serum-free Opti-Minimal Essential Medium I (without antibiotics; Invitrogen, Waltham, MA, USA) were diluted. The siRNA sequences used to transfect the siRHAMM (RHAMM knockdown) group were RHAMM-siRNA 1, RHAMM-siRNA 2, and RHAMM-siRNA 3. The siRNA sequence used for the negative control (NC) group was nonspecific siRNA. The same volume of Opti- Minimal Essential Medium I (instead of siRNA) was used in the control (mock transfection) group. Next, contents were mixed by carefully pipetting up and down, and the transfection reagent was then incubated with the siRNA complex for 15–20 min at approximately 15–20 °C. The siRNA complex was then applied dropwise to the cells and gently mixed to ensure even distribution over the entire plate surface. After 6 h of incubation, the medium was replaced with serum-containing medium. RHAMM mRNA expression was detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) at 24 and 48 h post-transfection. RHAMM protein expression was detected by western blot analysis at 48 h post-transfection.
RT-qPCR
Total RNA was extracted using the TRIzol method (Invitrogen), and RT-qPCR was performed using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan). RT-qPCR was conducted with the Go Taq® qPCR Master Mix kit (Promega Corporation, Madison, WI, USA), according to the manufacturer’s protocols. Thermocycling conditions were set as pre-denaturation at 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 1 min and elongation at 95 °C for 15 s. DNA amplification primer sequences (Invitrogen) were RHAMM (forward, 5'-TTG CCC TGA AGA CCC CAT T-3'; reverse, 5'-TGT TCC TTT CAC ATA TTT AAG GAT TG-3') and GAPDH (forward, 5'-GCA CCG TCA AGG CTG AGA AC-3'; reverse, 5'-TGG TGA AGA CGC CAG TGG A-3'). GAPDH served as an internal reference. Relative RHAMM mRNA expression was calculated using the 2−ΔΔCq method (19).
Western blot
Cells from six-well plates were washed twice with cold PBS, lysed in lysis buffer, and centrifuged at 12,000 ×g for 10 min at 4 °C for total protein isolation. Protein concentration was determined using a BCA protein assay kit (cat. no. WLA004; Wanleibio Co., Ltd., Shanghai, China). Samples were submitted to sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, blocked with 5% non-fat milk containing blocking solution at room temperature for 2 h, and then washed. The primary antibodies used were rabbit anti-CD168 antibody (EPR4055; cat. no. ab108339; dilution, 1:5,000; Abcam, Cambridge, UK), rabbit anti-GAPDH antibody (cat. no. WL03368; dilution, 1:1,000; Wanleibio Co., Ltd.) as a reference protein, rabbit anti- ERK1/2 antibody (cat. no. WL01864; dilution, 1:500; Wanleibio Co., Ltd.), rabbit anti-phosphorylated (p)-ERK1/2 antibody (cat. no. WL02368; dilution, 1:500; Wanleibio Co., Ltd.), and rabbit anti-β-actin antibody (cat. no. WL01845; dilution, 1:1,000; Wanleibio Co., Ltd.) as a reference protein. Primary antibodies were incubated with membranes overnight at 4˚C, and then incubated with horseradish peroxidase-labeled secondary antibody goat anti-rabbit IgG (cat. no. WLA023; dilution, 1:5,000; Wanleibio Co., Ltd.) for 45 min at 37 °C. Membranes were then washed and analyzed using an electrochemiluminescence kit (cat. no. WLA003; Wanleibio Co., Ltd.).
Irradiation
Cells were irradiated using a 6-MV X-ray linear accelerator (Varian, Palo Alto, CA, USA) with a tissue compensator of 15 mm, source skin distance of 100 cm, and dose rate of 400 cGy/min at room temperature.
Colony formation assay
A549 and H460 cells were seeded into six-well plates at different cell densities (200, 400, 800, 2,000, 5,000, and 10,000 cells/well; n=3). After one-night post-seeding, cells attached to plates were irradiated at various doses (0, 2, 4, 6, 8, and 10 Gy) and cultured continuously for 10–14 days. Next, cells were washed with PBS, fixed using 95% alcohol for 10 min, stained with crystal violet for 15 min at room temperature, and flushed with distilled water prior to colony counting. Experiments were repeated three times. Calculations were made as follows: colony formation rate = calculated number of cell colones/number of seeded cells; and cell surviving fraction = irradiated cell colony formation rate/control (0 Gy) cell colony formation rate. Using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA), the dose-survival curve was generated using a multi-target click model; surviving fraction=1− (1−e−kD)N. The k constant relates to ray quality and cell radiosensitivity.
Then, A549 cells were divided into three groups: Control + R (mock transfection plus radiation), NC + R (negative control plus radiation), and siRHAMM + R (RHAMM knockdown plus radiation). At 8 h post-transfection, cells were seeded into six-well plates, and experiments were conducted as described previously. The sensitivity enhancement ratio (SER) value was calculated as the k value of the siRHAMM group/k value of the Control group.
Cell proliferation
Cells were divided into six groups: Control, NC, siRHAMM, Control + R, NC + R, and siRHAMM + R. One day prior to transfection, cells were cultured overnight in 96-well plates at 1×104 cells/well. At 24 h post-transfection, cells in the three radiation groups were irradiated with 4 Gy, and cell counting was performed using Cell Counting Kit-8 assays (Tongren Chemical Company, Kumamoto Prefecture, Japan) at 24 and 48 h following irradiation. The absorbance [optical density (OD)] value was measured at 490 nm. The OD value indirectly reflected the number of living cells; Therefore, CCK-8 assays could be used to analyze cell proliferation.
Cell apoptosis
Cells were divided into six groups: Control, NC, siRHAMM, Control + R, NC + R, and siRHAMM + R. One day prior to transfection, cells were cultured overnight in 24-well plates at 4×104 cells/well. Cells in the three radiation groups were then irradiated with 4 Gy at 24 h post-transfection. At 24 h post-irradiation, single-cell suspensions were incubated with annexin V and fluorescein isothiocyanate (FITC)-labeled propidium iodide (PI) and placed on a flow cytometer. The FITC Annexin V Apoptosis Detection Kit I was purchased from BD Biosciences (Franklin Lakes, NJ, USA).
Cell cycle
Cells were divided into six groups: Control, NC, siRHAMM, Control + R, NC + R, and siRHAMM + R. One day prior to transfection, cells were cultured overnight in six-well plates at 2×105 cells/well. Cells in the three radiation groups were irradiated with 4 Gy at 24 h post-transfection. At 24 h post-irradiation, single-cell suspensions were fixed in 70% ethanol at 4 °C overnight, washed with cold PBS, and then incubated with RNase A at 37 °C in a water bath for 30 min. Next, cells were stained with PI at 4 °C in the dark for 30 min. The cell cycle was evaluated by flow cytometry using a cell cycle detection kit was purchased from Jiangsu KGI Biotechnology Co., Ltd (Jiangsu, China).
Expression of ERK1/2 and p-ERK1/2
Cells were divided into three groups: Control + R, NC + R, and siRHAMM + R. One day prior to transfection, cells were cultured overnight in six-well plates at 2×105 cells/well, and cells were irradiated with 4 Gy at 24 h post-transfection. ERK1/2 and p-ERK1/2 expression was detected by western blot at 24 h post-irradiation.
Statistical analysis
For all assays, experiments were repeated at least three times. Values are expressed as means ± standard deviations. Using SPSS 11.5 software (SPSS, Inc., Chicago, IL, USA) differences between groups were evaluated by one-way analysis of variance, followed by Tukey’s post hoc test. P values <0.05 were considered to be statistically significant.
Results
RHAMM gene expression negatively correlated with radiosensitivity of NSCLC cell lines
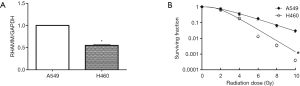
RT-qPCR was used to detect RHAMM gene expression in A549 and H460 cell lines. Assuming that the expression level of the A549 cell line was 1.00, the relative expression level of the H460 cell line was 0.55±0.02 (P<0.05; Figure 1A). Colony formation assays showed that radiosensitivity of the A549 cell line was significantly weaker than that of the H460 cell line (P<0.05; Figure 1B). We considered that RHAMM gene expression was negatively correlated with the radiosensitivity of NSCLC cell lines.
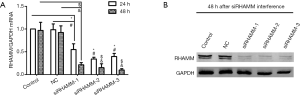
RHAMM gene silencing
To assess its effect on A549 cells, RHAMM expression was silenced by siRNA and evaluated by RT-qPCR and western blot analysis following transfection. RHAMM mRNA levels decreased at 24 and 48 h following RHAMM siRNA transfection in the siRHAMM group compared with the Control and NC groups (P<0.05; Figure 2A). Expression of RHAMM protein 48 h post-transfection was also reduced (Figure 2B). These results suggest that siRNA transfection inhibited RHAMM expression in A549 cells. The siRNA sequence RHAMM-siRNA 2 was used in subsequent experiments; this sequence reduced RHAMM expression at the mRNA and protein levels by 84% and 93%, respectively, at 48 h post-transfection.
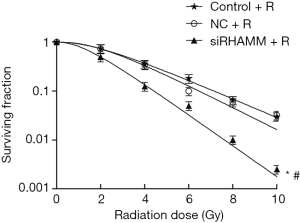
Radiosensitivity of A549 cells was enhanced following RHAMM gene silencing
Colony formation assays were conducted to determine the radiosensitivity effect of RHAMM on A549 cells. Radiosensitivity in the siRHAMM + R group was enhanced (P<0.05), and the survival curve shifted to the left, resulting in a SER of 1.63 (Figure 3). This result indicated that RHAMM had an inhibitory effect on A549 cell radiosensitivity.
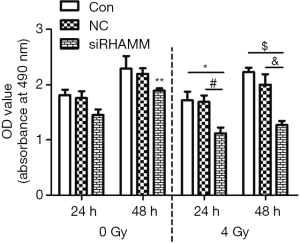
A549 cell proliferation decreased following RHAMM gene silencing plus radiation
A549 cell proliferation was detected with the Cell Counting Kit-8. At 24 h post-transfection, cells in the three radiation groups were irradiated with 4 Gy, and proliferation levels were determined at 24 and 48 h following irradiation. At 24 and 48 h, OD values were lower in the siRHAMM + R group (1.12±0.18 and 1.27±0.16, respectively) than in the Control + R group (1.72±0.27 and 2.23±0.18, respectively) and NC + R group (1.69±0.24 and 2.00±0.42, respectively); At 48 h, the OD value was lower in the siRHAMM + R group than in the siRHAMM group (1.89±0.09; P<0.05; Figure 4). These results shown that less A549 cell proliferation occurred in the siRHAMM + R group than in the siRHAMM and Control + R groups.
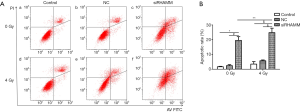
Radiation-induced cell apoptosis increased following RHAMM gene silencing
The rate of apoptosis was evaluated using Annexin V/PI staining and a flow cytometer. A549 cells were divided into irradiation and non-radiation groups. The irradiation group was exposed to 4 Gy at 24 h post-transfection. At 48 h post-transfection, the apoptotic rate was determined for all groups. The apoptotic rate was higher in the siRHAMM group (19.57%±2.51%) than in the Control (1.67%±0.30%; P<0.05) and NC (2.42%±0.66%; P<0.05) groups. The apoptotic rate was higher in the siRHAMM + R group (24.81%±3.05%) than in the siRHAMM, Control + R (3.45%±1.73%; P<0.05) and NC + R (5.71%±0.58%; P<0.05) groups (Figure 5). Knockdown of RHAMM increased the apoptotic rate, and an additional apoptotic effect was observed in the RHAMM knockdown plus radiation group. Thus, RHAMM gene silencing increased radiation- induced apoptosis.
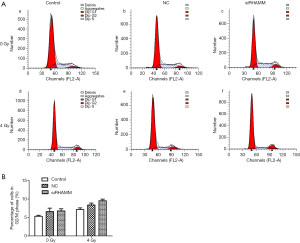
Percentage of cells in the G2/M phase was unchanged following RHAMM gene silencing
Cell cycle changes following RHAMM gene silencing and irradiation were evaluated by flow cytometric analysis. The ratio of cells in the G2/M phase did not differ between the siRHAMM group (6.78%±0.94%) and the Control and NC groups (5.33%±0.48% and 6.6%±1.57%, respectively). Similarly, the ratio of cells in the G2/M phase did not differ between the siRHAMM + R group (9.51%±0.76%) and the Control + R and NC + R groups (7.16%±0.72% and 8.38%±0.89%, respectively) (Figure 6).
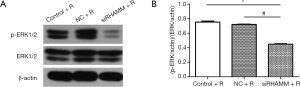
ERK1/2 phosphorylation was reduced following RHAMM gene silencing
The present study also aimed to determine which signaling pathway RHAMM used to inhibit the radiosensitivity of A549 cells; therefore, ERK1/2 and p-ERK1/2 protein expression was measured by western blot. Although p-ERK1/2 expression was lower in the siRHAMM + R group than in the Control + R and NC + R groups, ERK1/2 expression did not differ between groups. The ratio of (p-ERK/actin)/(ERK/actin) was lower in the siRHAMM + R group (0.45±0.01) than in the Control + R (0.75±0.02) and NC + R (0.72±0.01) groups (P<0.05 for both; Figure 7). Thus, RHAMM gene silencing reduced ERK1/2 phosphorylation and regulated the ERK1/2 signaling pathway.
Discussion
The effect of radiation on cancer cells is heterogeneous and complex due to a number of factors, including race, region, origin, and pathogenicity. The intrinsic radiosensitivity of cancer cells has been shown to be related to level of hypoxia, DNA repair capacity, numbers of dividing and apoptotic cells and cell cycle phases (1), and genotype-dependent radiosensitivity pattern (20).
First, we determined that RHAMM gene expression in the NSCLC cell line A549 was higher than that in cell line H460. Next, we wanted to understand the relationship between RHAMM gene expression and radiosensitivity of the two cell lines. We hypothesized that RHAMM gene expression was negatively correlated with NSCLC cell line radiosensitivity.
Could RHAMM gene inhibition enhance radiosensitivity? In the present study, inhibition of A549 RHAMM gene expression increased cancer cell radiosensitivity. This finding shows that targeting of the RHAMM gene in vitro can enhance NSCLC radiosensitivity.
RHAMM is highly expressed in tumors with muscle invasion, high-grade tumors, tumors with lymph node involvement, and recurrent tumors (8). In addition to increased overall expression, RHAMM subtype expression varies, with some subtypes adopting the role of multifunctional oncogenes. Oncogenes serve an important role in the signal transduction pathway of cell growth and differentiation. Nuclear factor-κB (NF-κB), which can be activated by radiation, binds directly to the human epidermal growth factor receptor 2 (HER2) promoter to cause HER2 overexpression and tumor radioresistance. Additionally, HER2 can activate basal and radiation-induced NF-κB activity through activation of the phosphoinositide 3-kinase/protein kinase B pathway to further induce HER2 overexpression. Targeting of the NF-κB/HER2 pathway in radiation-resistant recurrent tumors may enhance breast cancer cure rates (21). Much evidence suggests that the epithelial growth factor receptor (EGFR) signaling pathway is important in the tumor cell response to irradiation. EGFR signaling is mediated by mitogen extracellular kinase-extracellular signal-regulated kinase and phosphoinositide 3-kinase/protein kinase B in the response of glioblastoma multiforme spheroids to radiation (22). A clinical EGFR inhibitor combined with radiotherapy can increase tumor killing and improve local control (23). RHAMM may serve as an oncogene to inhibit the radiosensitivity of lung adenocarcinoma A549 cells; thus, radiosensitivity may be enhanced through RHAMM gene silencing.
Schütze and colleagues (16) reported that neither irradiation nor siRNA transfection changed the proliferation rate in human breast cancer cell lines. In this study, RHAMM gene expression was inhibited in the presence of radiotherapy, and A549 cell proliferation decreased. The mechanism by which intracellular or extracellular RHAMM promotes tumorigenesis and progression remains unclear (24). RHAMM was originally discovered as an HA-binding protein that was secreted into the extracellular matrix to activate cell-surface transmembrane adhesion molecules and HA receptor CD44 for cell migration and invasion. This RHAMM-regulated activation increases CD44 expression on cell surfaces and in CD44-mediated ERK1/2 activation (25,26). The strong association between the RHAMM-CD44 and RAS/ERK1/2 signaling pathways suggests that these HA receptor complexes are able to control signaling pathways that are overactivated in many tumor types (27). Additionally, cell surface RHAMM also interacts with recepteur d’origine nantais, a receptor tyrosine kinase of the c-Met proto-oncogene family (28). Thus, extracellular RHAMM interacts with one or more adhesion/growth factor receptors on the cell surface to activate key signaling pathways and promote tumor metastasis. Intracellular RHAMM, which has been observed in human breast cancer cells, regulates cytoskeletal organization via interactions with microtubules and actin filaments (29) and contributes to the activation of ERK (17,30,31).
The results from the present study also showed that the apoptotic rate of A549 cells was increased following inhibition of all splice variants of RHAMM, and that the apoptotic rate in A549 cells was further increased following gene suppression and radiotherapy. Schütze and colleagues (16) addressed the potential role of short interfering RNAs against RHAMM and its splice variants. They found that RHAMMv1/v2 knock-down in RHAMMhigh MCF-7 cells, but not RHAMMlow MDA-MB-231 cells, increased the apoptotic rate after irradiation. Apoptosis is the main method of cell death following radiation exposure. Radiosensitive tumors have high levels of apoptosis following radiotherapy, and apoptosis response can be used to predict a tumor’s inherent radiosensitivity (32). The balance between apoptosis and anti-apoptosis signaling is achieved through the regulation of various factors (e.g., apoptosis-related genes). This regulation has been a key area of recent research interest.
The present study explored cell cycle changes in response to RHAMM gene inhibition. Cells in the G2/M phase did not increase following gene silencing or gene silencing combined with radiotherapy. Although the radioresistant phenotype can be changed to a radiosensitive phenotype during fractionated radiotherapy, failure of the cell cycle to change could lead to radioresistance (33). Radiosensitivity varies by cell cycle. For example, cells are radioresistant in S, relatively radiosensitive in G0/G1, and radiosensitive in G2/M (34). Ionizing radiation can induce changes in cell cycle progression, such as G1 and G2 phase arrest. Arrest time depends on a cell’s intrinsic radiosensitivity, which is the key event by which cells verify the authenticity and integrity of hereditary substances and repair damage. However, the detailed molecular mechanisms for this process remain unknown (35).
The present study showed that irradiated A549 cells had significantly reduced ERK1/2 phosphorylation due to the inhibition of RHAMM expression. Aberrant ERK1/2 signaling in tumor cells leads to enhanced or abnormal cell proliferation, antagonism of apoptosis, or antagonism of chemoradiation and targeted therapy. Previous studies have demonstrated that low-dose radiation can protect cells from radiation-induced damage by promoting cell growth and proliferation; this process is closely associated with ERK1/2 activation in normal and tumor cells (36,37). Recent, studies have shown that ERK1/2 activation in many types of tumor cell contributes to radioresistance (38,39). Thus, we can infer that the inhibition of RHAMM expression combined with radiotherapy reduces ERK1/2 activation to enhance the radiosensitivity of A549 cells.
In conclusion, RHAMM gene expression was negatively correlated with the radiosensitivity of NSCLC cell lines. Inhibition of RHAMM gene expression enhanced the radiosensitivity of lung adenocarcinoma cell line A549, reduced proliferation by enhancing radiosensitivity through an increase in the apoptotic rate, reduced ERK1/2 phosphorylation, and regulated the ERK1/2 signaling pathway. Therefore, RHAMM is a potential target for radiosensitization. These results may lead to novel approaches to gene therapy and radiosensitization of lung cancer. However, more research is needed to understand the role of RHAMM in DNA damage repair, determine whether this target gene can be used in vivo, and determine the effect of this radiosensitization method on normal tissue.
Acknowledgments
We thank Professor Yue Zhang from the Rheumatology Laboratory and radiation oncology technicians from the First Affiliated Hospital of Harbin Medical University for technical assistance.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.02.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional Review Board approval was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cao R, Ding Q, Li P, et al. SHP1-mediated cell cycle redistribution inhibits radiosensitivity of non-small cell lung cancer. Radiat Oncol 2013;8:178. [Crossref] [PubMed]
- Wirsdörfer F, Cappuccini F, Niazman M, et al. Thorax irradiation triggers a local and systemic accumulation of immunosuppressive CD4+ FoxP3+ regulatory T cells. Radiat Oncol 2014;9:98-108. [Crossref] [PubMed]
- Wang C, Entwistle J, Hou G, et al. The characterization of a human RHAMMcDNA: conservation of the hyaluronan-binding domains. Gene 1996;174:299-306. [Crossref] [PubMed]
- Du YC, Chou CK, Klimstra DS, et al. Receptor for hyaluronan-mediated motility isoform B promotes liver metastasis in a mouse model of multistep tumorigenesis and a tail vein assay for metastasis. Proc Natl Acad Sci 2011;108:16753-8. [Crossref] [PubMed]
- Greiner J, Ringhoffer M, Taniguchi M, et al. Receptor for hyaluronanacid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol 2002;30:1029-35. [Crossref] [PubMed]
- Li H, Guo L, Li JW, et al. Expression of hyaluronan receptors CD44 and RHAMM in stomach cancers: relevance with tumor progression. Int J Oncol 2000;17:927-32. [PubMed]
- Rein DT, Roehrig K, Schondorf T, et al. Expression of the hyaluronan receptor RHAMM in endometrial carcinoma ssuggests a role in tumour progression and metastasis. J Cancer Res Clin Oncol 2003;129:161-4. [PubMed]
- Niedworok C, Kretschmer I, Röck K, et al. The impact of the receptor of hyaluronan-mediated motility (RHAMM) on human urothelial transitional cell cancer of the bladder. PLoS One 2013;8:e75681. [Crossref] [PubMed]
- Rizzardi AE, Rosener NK, Koopmeiners JS, et al. Evaluation of protein biomarkers of prostate cancer aggressiveness. BMC Cancer 2014;14:244. [Crossref] [PubMed]
- Venables JP, Klinck R, Bramard A, et al. Identifcation of alternative splicing markers for breast cancer. Cancer Res 2008;68:9525-31. [Crossref] [PubMed]
- Schmitt A, Barth TF, Beyer E, et al. The tumor antigens RHAMM and G250/CAIX are expressed in head and neck squamous cell carcinomas and elicit specific CD8+ T cell responses. Int J Oncol 2009;34:629-39. [Crossref] [PubMed]
- Stangeland B, Mughal AA, Grieg Z, et al. Combined expressional analysis, bioinformatics and targeted proteomicsidentify new potential therapeutic targets in glioblastoma stem cells. Oncotarget 2015;6:26192-215. [Crossref] [PubMed]
- Wang D, Narula N, Azzopardi S, et al. Expression of the receptor for hyaluronic acid mediated motility (RHAMM) is associated with poor prognosis and metastasis in non-small cell lung carcinoma. Oncotarget 2016;7:39957-69. [PubMed]
- Augustin F, Fiegl M, Schmid T, et al. Receptor for hyaluronic acid-mediated motility (RHAMM, CD168) expression is prognostically importantin both nodal negative and nodal positive large cell lung cancer. J Clin Pathol 2015;68:368-73. [Crossref] [PubMed]
- Thangavel C, Boopathi E, Liu Y, et al. RB Loss Promotes Prostate Cancer Metastasis. Cancer Res 2017;77:982-95. [Crossref] [PubMed]
- Schütze A, Vogeley C, Gorges T, et al. RHAMM splice variants confer radiosensitivity in human breast cancer cell lines. Oncotarget 2016;7:21428-40. [Crossref] [PubMed]
- Hatano H, Shigeishi H, Kudo Y, et al. RHAMM/ERK interaction induces proliferative activities of cementifying fibroma cells through a mechanism based on the CD44-EGFR. Lab Invest 2011;91:379-91. [Crossref] [PubMed]
- Steinmetz R, Wagoner HA, Zeng P, et al. Mechanisms regulating the constitutive activation of the extracellular signal-regulated kinase (ERK) signaling pathway in ovarian cancer and the effect of ribonucleic acid interference for ERK1/2 on cancer cell proliferation. Mol Endocrinol 2004;18:2570-82. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-∆∆ C(T)) method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Williams JR, Zhang Y, Zhou H, et al. A quantitative overview of radiosensitivity of human tumor cells across histological type and TP53 status. Int J Radiat Biol 2008;84:253-64. [Crossref] [PubMed]
- Cao N, Li S, Wang Z, et al. NF-κB-Mediated HER2 Overexpression in Radiation-Adaptive Resistance. Radiat Res 2009;171:9-21. [Crossref] [PubMed]
- Fedrigo CA, Grivicich I, Schunemann DP, et al. Radioresistance of human glioma spheroids and expression of HSP70, p53 and EGFr. Radiat Oncol 2011;6:156-65. [Crossref] [PubMed]
- Bergkvist GT, Argyle DJ, Pang LY, et al. Studies on the inhibition of feline EGFR in squamous cell carcinoma: Enhancement of radiosensitivity and rescue of resistance to small molecule inhibitors. Cancer Biol Ther 2011;11:927-37. [Crossref] [PubMed]
- Telmer PG, Tolg C, McCarthy JB, et al. How does a protein with dual mitotic spindle and extracellular matrix receptor functions affect tumor susceptibility and progression? Commun Integr Biol 2011;4:182-5. [Crossref] [PubMed]
- Tolg C, Hamilton SR, Nakrieko KA, et al. Rhamm-/- fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound. J Cell Biol 2006;175:1017-28. [Crossref] [PubMed]
- Hamilton SR, Fard SF, Paiwand FF, et al. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1, 2 that sustain high basal motility in breast cancer cells. J Biol Chem 2007;282:16667-80. [Crossref] [PubMed]
- Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer 2006;6:735-41. [Crossref] [PubMed]
- Manzanares D, Monzon ME, Savani RC, et al. Apical oxidative hyaluronan degradation stimulates airway ciliary beating via RHAMM and RON. Am J Respir Cell Mol Biol 2007;37:160-8. [Crossref] [PubMed]
- Assmann V, Jenkinson D, Marshall JF, et al. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin flaments. J Cell Sci 1999;112:3943-54. [PubMed]
- Zhang S, Chang MC, Zylka D, et al. The hyaluronan receptor RHAMM regulates extracellular regulated kinase. J Biol Chem 1998;273:11342-8. [Crossref] [PubMed]
- Wang Z, Wu Y, Wang H, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci USA 2014;111:E89-98. [Crossref] [PubMed]
- Decaudin D, Delic J, Dumont J, et al. Clinical effcacy of irradiation in CLL patients: predictive value of
in vitro radio-induced apoptosis. Leuk. Lymphoma 2002;43:827-9. [Crossref] [PubMed] - Cao R, Ding Q, Li P, et al. SHP1-mediated cell cycle redistribution inhibits radiosensitivity of non-small cell lung cancer. Radiat Oncol 2013;8:178-86. [Crossref] [PubMed]
- Teyssier F, Bay JO, Dionet C, et al. Cell cycle regulation after exposure to ionizing radiation. Bull Cancer 1999;86:345-57. [PubMed]
- Lieberman HB, Hopkins KM. Schizosaccharomyces malidevorans and Sz. octosporus homologues of Sz. pombe rad9, a gene that mediates radioresistance and cell-cycle progression. Gene 1994;150:281-6. [Crossref] [PubMed]
- Drigotas M, Affolter A, Mann WJ, et al. Reactive oxygen species activation of MAPK pathway results in VEGF upregulation as an undesired irradiation response. J Oral Pathol Med 2013;42:612-9. [Crossref] [PubMed]
- Dent P, Yacoub A, Fisher PB, et al. MAPK pathways in radiation responses. Oncogene 2003;22:5885-96. [Crossref] [PubMed]
- Kotowski U, Heiduschka G, Brunner M, et al. Radiosensitization of Head and Neck Cancer Cells by the Phytochemical Agent Sulforaphane. Strahlenther Onkol 2011;187:575-80. [Crossref] [PubMed]
- Carón RW, Yacoub A, Mitchell C, et al. Radiationstimulated ERK1/2 and JNK1/2 signaling can promote cell cycle progression in human colon cancer cells. Cell Cycle 2005;4:456-64. [Crossref] [PubMed]


