The proteomic comparison of peripheral circulation-derived exosomes from the epithelial ovarian carcinoma (EOC) patients and non-EOC subjects
Introduction
Epithelial ovarian carcinoma (EOC), for which screening and diagnostic methods are suboptimal, is the most lethal gynecological cancer (1). Unfortunately, current diagnostic methods, such as serum cancer antigen 125 (CA125) detection and pelvic ultrasonic scanning (2), lack sensitivity and specificity, and computed tomography, magnetic resonance imaging and positron emission tomography are relatively expensive for use as an early screening or diagnostic tool. Therefore, development of a strategy to screen or diagnose EOC as early as possible is important for improving the prognosis of EOC.
Exosomes are nanoscale biovesicles secreted by numerous types of cells, including immune cells (3,4), red blood cells (4) and tumor cells (5). Typically, exosomes have a cup-like shape with a diameter of 30–100 nm. Because exosomes can carry cellular components such as proteins (6,7), miRNAs (7) and mRNAs (8) from donor cells, which may indicate the donor cells’ biological functions or conditions, exosomes are regarded as cell-to-cell messengers, and their cargoes may be potential tumor markers for screening or diagnosing tumors. In addition to tumor markers, the cargoes of exosomes or extracellular vesicles (EVs) are able to influence the host-tumor interaction, such as inhibition of T cell anti-tumor activity (9). Recently, ovarian cancer cells were shown to secrete exosomes or EVs into the outer cellular microenvironment or space, such as the peripheral circulation (10) and ascites (9,11); thus, tumor-derived exosomes, particularly the molecular components loaded in exosomes, might be used as a noninvasive marker for early detection of EOC.
In a previous study (12), we analyzed the protein components loaded into tumor-derived exosomes from two ovarian cancer lines. To further elucidate the biological characteristics of EOC and explore a method for early detection of EOC, in addition to identifying proteins that may be potential tumor markers, we designed this study to compare the different proteins loaded in exosomes in vivo from EOC patients and non-EOC subjects and to verify the in vivo results by using the in vitro results from ovarian cancer cell lines.
Methods
Patient and control populations
From January 1st, 2015 to December 31st, 2015, 10 patients with EOC (cancer group), primarily diagnosed by diagnostic laparoscopy or exploratory laparotomy in our department, were recruited, and a gynecological pathologist at our hospital confirmed the diagnosis of EOC. All EOC patients had the serous carcinoma subtype. Among the patients, 8 were classified as International Federation of Gynecology and Obstetrics (FIGO) stage IIB; 1 as stage IA; and 1 as stage IC2. The non-EOC subjects (control group) consisted of 10 women recruited from patients with pelvic floor dysfunction (PFD) who required surgical treatment in the same period as the cancer patients in our department. Before enrollment in the study, the control group subjects received a physical examination to exclude the possibility of any tumors.
Under the rules of the Ethical Committee of our institution, blood sample collection does not require the approval of the committee but must be permitted by the patient. Therefore, before recruitment, all subjects provided signed informed consent and approved their blood samples for use by the researchers.
Collection of ovarian cancer tissues and normal ovarian tissues
A 2×2 cm sample of fresh ovarian epithelial cancer tissue was obtained through diagnostic laparoscopy, exploratory laparotomy or cytoreductive surgery and immediately stored in liquid nitrogen for further experiments. The same volume of fresh normal ovarian tissue was obtained from the PFD patients who received hysterectomy plus oophorectomy, and the ovarian tissue was immediately stored in liquid nitrogen in the operation room.
Isolation and identification of exosomes from the peripheral circulation
Isolation and purification of exosomes
Before the blood samples were obtained, EOC patients had never received any tumor-associated treatment, such as debulking surgery, chemotherapy, radiotherapy or targeted therapy. First, 5 mL peripheral blood samples from every EOC patient or control subject were obtained, and serum was isolated via centrifugation at 10 min at 4 °C. Exosomes were isolated as described in our previous study (13). Briefly, the serum was centrifuged once at 500 g for 10 min and then at 2,000 g for 20 min to eliminate cell and cell debris contamination. The supernatant was further centrifuged at 16,500 g for 30 min at 4 °C. Then, the exosomes were pelleted by ultracentrifugation at 120,000 g for 80 min at 4 °C. The exosome pellets were subsequently washed once in phosphate-buffered saline (PBS) and pelleted by ultracentrifugation at 120,000 g for 80 min at 4 °C. The final exosome pellets were resuspended in PBS and filtered through a 0.22 µm membrane. The exosomes were stored at −80 °C for further analysis.
Identification of exosomes by transmission electron microscopy
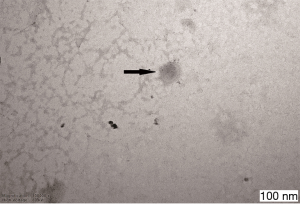
The obtained exosomes were pooled and loaded onto Formvar carbon-coated 200 mesh copper grids and allowed to absorb at room temperature (RT) for 10 min. The absorbed exosomes were negatively stained with 3% phosphotungstic acid at RT for 5 min. The exosome-containing grids were air-dried and observed with a Hitachi H-7600 transmission electron microscope operating at 80.0 kV.
Nanoparticle tracking analysis (NTA) of exosomes
Exosomes from cancer serum and control serum, suspended in 1 mL of PBS, were injected into the NTA system (ZETASIZER Nanoseries-Nano-ZS, Malvern, UK). Then, the diameter distribution of nanoparticles was tested as a manipulation guide.
Western blot analysis of exosomes
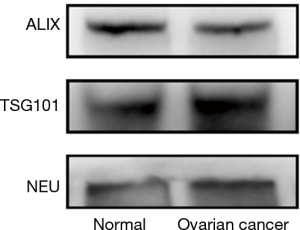
Exosomes were prepared as described above. The samples were pooled for subsequent western blotting. First, 5× SDS-loading buffer was added to dissolve the exosome proteins, and the samples were then heated at 95 °C for 5 min. The samples were next centrifuged at 13,000 rpm for 5 min to remove insoluble materials. The supernatants were subsequently separated through SDS-polyacrylamide gel electrophoresis (SDS-PAGE, 3% stacking gel, 12–15% running gel) in a Mini Protean 2 electrophoresis system (Bio-Rad, Munich, Germany). The proteins were transferred to a polyvinylidene fluoride (PVDF) membrane in transfer buffer. After being blocked with 5% nonfat milk in PBS with 0.5% Tween-20 (PBST) for 1 hour at RT, the membrane was incubated with the primary antibody (TSG101 Abcam, No 125011, USA, Alix, Abcam, No 186429, USA and Neu, Abcam, No 214275, USA) overnight at 4 °C. Immunocomplexes were labeled with an HRP-conjugated secondary antibody and detected using an enhanced chemiluminescence (ECL, Bio-Rad, Munich, Germany) system. Between each incubation step, the membrane was washed three times with 0.5% PBST.
Proteomic analyses of exosomes, ovarian cancer tissues and normal ovarian tissues
Extraction of proteins from ovarian cancer or normal ovarian tissues
Ovarian cancer and normal ovarian tissues were ground in liquid nitrogen and then precipitated in trichloroacetic acid and acetone at −20 °C for 2 hours. Then, the proteins were centrifuged at 20,000 g for 30 min at 4 °C, and the sediments were washed with cold acetone and reprecipitated at −20 °C for 30 min, followed by centrifugation at 20,000 g for 30 min at 4 °C. The procedure was repeated several times if necessary. Finally, the protein sediments were lysed with lysis buffer under ultrasonic conditions.
Extraction of proteins from exosomes
First, the protein components were precipitated via the method described above but were not lysed in lysis buffer. Second, dithiothreitol was added to the protein suspension, followed by incubation in a water bath at 56 °C for 1 hour. Then, 2-iodoacetamide was immediately added to the protein solution, followed by incubation in a dark room for 1 hour. The proteins were precipitated using cold acetone at −20 °C, followed by centrifugation at 20,000 g for 30 min at 4 °C. Finally, the sediment from the last step was dissolved in lysis buffer and recentrifuged as in the above step, and the suspension was then stored at −80 °C for further analysis.
Quantification and labeling of proteins
The protein concentration was determined with the Bradford method using bovine serum albumin as a standard. After the samples were digested by trypsin, 100 µg protein samples from ovarian cancer tissues, normal ovarian tissues, patient-derived exosomes and volunteer-derived exosomes were labeled following the manufacturer’s instructions for the isobaric tags for relative and absolute quantitation (iTRAQ) system (Applied Biosystems, MA, USA). The labeled samples were pooled for high-performance liquid chromatography/mass spectrometry (HPLC/MS) analysis.
HPLC/MS analyses of exosomes and ovarian tissues
The extracted peptides from the 100 µg protein samples were separated in an analytical capillary column (50 µm × 10 cm) packed with 5 µm spherical C18 reversed-phase material (YMC, Kyoto, Japan). A DIONEX U3000 nanoRSLC HPLC system (Dionex, CA, USA) was used to generate the HPLC gradient of 0–30% B for 40 min and 30–80% B for 15 min (A =0.1% formic acid in water, B =0.1% formic acid in acetonitrile). The eluted peptides were sprayed into a Q-Exactive mass spectrometer (Thermo Fisher Scientific, CA, USA). The mass spectrometer was operated in positive ion mode with one MS scan followed by ten high-energy collisional dissociation MS/MS scans for each cycle.
Database searches were performed using Proteome Discoverer (version 1.3, Thermo Fisher Scientific, CA, USA) and Mascot (version 2.3.0, Matrix Science, Ltd., London, UK). The search parameters were as follows: 15 ppm mass tolerance for precursor ions and one missed cleavage site were allowed for trypsin digestion. The search results were filtered with both the peptide significance threshold and expectation value below 0.05. The Mascot Percolator scores were used for all peptides.
Western blot analysis of the results from HPLC/MS analyses
The proteomic results from HPLC/MS were confirmed by western blotting. Samples of 5 µg of total exosomal proteins from both the cancer group and the control group were used for testing. The following antibodies were used for western blotting: CLIC4 (Abcam, No183043, UK), ATK1 (Abcam, No89402, UK), EPAP II (Abcam, No188320, UK), SNX3 (Abcam, No56078, UK), FAM49B (Abcam, No121299, UK), KIND3 (Abcam, No68040, UK), LTF (Abcam, No10110, UK) and TUBB3 (Abcam, No52623, UK).
Gene ontology (GO) annotation
To determine the subcellular localization of the identified proteins, we searched the LocDB (http://www.rostlab.org/services/locDB) database. Biological processes, molecular functions and protein classes and the pathways of the proteins involved were analyzed with the PANTHER (http://www.pantherdb.org/) consortium databases.
Exosomal protein database search
To confirm that the proteins identified in our study were found in other original exosomes, we searched the exosomal and EV molecular database Vesiclepedia (http://microvesicles.org/index.html).
Results
Characterization of exosomes from sera from EOC patients and control subjects
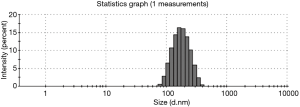
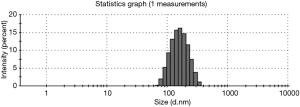
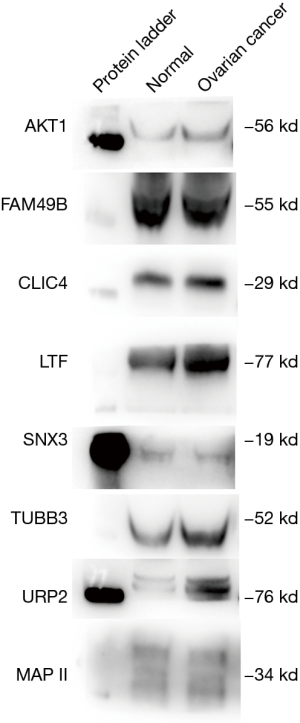
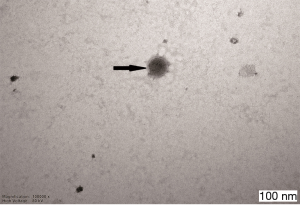
The exosomes from the cancer group and control group sera were isolated through a combination of differential centrifugation and ultrafiltration after removal of cell debris, nonmembrane proteins, protein aggregates and other larger vesicles. Under transmission electron microscopy, the preparations were shown to be mostly 30–180 nm in diameter (Figures 1,2). NTA data showed that the mean diameters of cancer derived-exosomes and control derived-exosomes were 158.6 and 148.7 nm, retrospectively (Figures 3,4). Western blot analyses detected the published exosomal markers TSG101 and Alix in exosomes from both groups, and the tumor marker Neu could be detected in the cancer group (Figure 5).
Proteomic analysis of exosomes derived from peripheral sera of ovarian cancer patients and control subjects
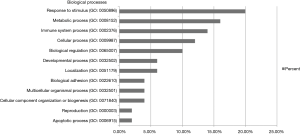
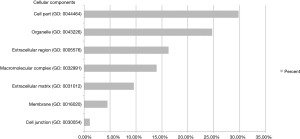
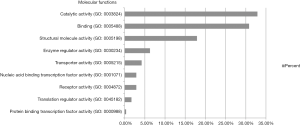
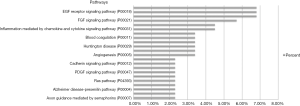
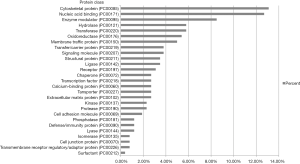
Protein database searching of HPLC/MS data identified 408 differentially expressed proteins in EOC and control exosomes, among which 208 proteins were downregulated, and 200 proteins were upregulated in EOC patient exosomes. The GO analysis results for the 200 upregulated proteins are shown in Figures 6-10, and GO enrichment analysis results are shown in Figure S1.
Proteomic analysis of proteins derived from ovarian cancer tissues and normal ovarian tissues
We identified 954 differentially expressed proteins in ovarian cancer tissues and normal ovarian tissues using the same technology that we used to explore the differentially expressed proteins in exosomes from different groups. There were 480 downregulated proteins and 474 upregulated proteins in the ovarian cancer tissues. We found that 35 proteins were upregulated in both ovarian cancer patient serum exosomes and ovarian cancer tissues (Table 1). Among these 35 proteins, 33 proteins were detected in the ExoCarta/Vesiclepedia database, and 15 of them were identified in the ovarian cancer cell lines OVCAR-1/IGROV-1 in our previous study (12) and ExoCarta/Vesiclepedia (Table 1). Interestingly, when compared with Hurwitz et al.’s study (14), we found that chloride intracellular channel protein 4 (CLIC4), serine/threonine-protein kinase 1 (AKT1), aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (EMAP II), sorting nexin-3 (SNX3), protein FAM49B (FAM49B), fermitin family homolog 3 (KIND3, also known as URP2), tubulin beta-3 chain (TUBB3) and lactotransferrin (LTF) were detected in this study, our previous study (12) and that of Hurwitz et al.
Table 1
| Protein description | Detected on ovarian cancer cell lines OVCAR3 and IGROV1 | Recorded on ExoCarta/Vesiclepedia database |
|---|---|---|
| Chloride intracellular channel protein 4 OS=Homo sapiens GN=CLIC4 PE=1 SV=4 - [CLIC4_HUMAN] | Yes | Yes |
| Coatomer subunit gamma OS=Homo sapiens GN=COPG PE=1 SV=1 - [COPG_HUMAN] | Yes | Yes |
| RNA-binding protein 8A OS=Homo sapiens GN=RBM8A PE=1 SV=1 - [RBM8A_HUMAN] | Yes | Yes |
| 7-dehydrocholesterol reductase OS=Homo sapiens GN=DHCR7 PE=1 SV=1 - [DHCR7_HUMAN] | Yes | Yes |
| Acid ceramidase OS=Homo sapiens GN=ASAH1 PE=1 SV=5 - [ASAH1_HUMAN] | Yes | Yes |
| Serine/threonine-protein kinase PAK 1 OS=Homo sapiens GN=PAK1 PE=1 SV=2 - [PAK1_HUMAN] | Yes | Yes |
| Aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 OS=Homo sapiens GN=AIMP1 PE=1 SV=2 - [AIMP1_HUMAN] | Yes | Yes |
| Sorting nexin-3 OS=Homo sapiens GN=SNX3 PE=1 SV=3 - [SNX3_HUMAN] | Yes | Yes |
| Protein FAM49B OS=Homo sapiens GN=FAM49B PE=1 SV=1 - [FA49B_HUMAN] | Yes | Yes |
| Tubulin beta-1 chain OS=Homo sapiens GN=TUBB1 PE=1 SV=1 - [TBB1_HUMAN] | Yes | Yes |
| Coronin-1B OS=Homo sapiens GN=CORO1B PE=1 SV=1 - [COR1B_HUMAN] | Yes | Yes |
| Fermitin family homolog 3 OS=Homo sapiens GN=FERMT3 PE=1 SV=1 - [URP2_HUMAN] | Yes | Yes |
| Tubulin beta-3 chain OS=Homo sapiens GN=TUBB3 PE=1 SV=2 - [TBB3_HUMAN] | Yes | Yes |
| Solute carrier family 2, facilitated glucose transporter member 1 OS=Homo sapiens GN=SLC2A1 PE=1 SV=2 - [GTR1_HUMAN] | Yes | Yes |
| Lactotransferrin OS=Homo sapiens GN=LTF PE=1 SV=6 - [TRFL_HUMAN] | Yes | Yes |
| tRNA methyltransferase 112 homolog OS=Homo sapiens GN=TRMT112 PE=1 SV=1 - [TR112_HUMAN] | No | Yes |
| Thioredoxin-related transmembrane protein 1 OS=Homo sapiens GN=TMX1 PE=1 SV=1 - [TMX1_HUMAN] | No | Yes |
| Thioredoxin domain-containing protein 17 OS=Homo sapiens GN=TXNDC17 PE=1 SV=1 - [TXD17_HUMAN] | No | Yes |
| E3 ubiquitin-protein ligase UHRF1 OS=Homo sapiens GN=UHRF1 PE=1 SV=1 - [UHRF1_HUMAN] | No | Yes |
| Reticulocalbin-3 OS=Homo sapiens GN=RCN3 PE=1 SV=1 - [RCN3_HUMAN] | No | Yes |
| Collagen triple helix repeat-containing protein 1 OS=Homo sapiens GN=CTHRC1 PE=1 SV=1 - [CTHR1_HUMAN] | No | Yes |
| Non-specific lipid-transfer protein OS=Homo sapiens GN=SCP2 PE=1 SV=2 - [NLTP_HUMAN] | No | Yes |
| Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 - [CAP7_HUMAN] | No | Yes |
| Protein S100-A9 OS=Homo sapiens GN=S100A9 PE=1 SV=1 - [S10A9_HUMAN] | No | Yes |
| Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 - [FIBG_HUMAN] | No | Yes |
| Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 - [FIBA_HUMAN] | No | Yes |
| Keratin, type II cytoskeletal 6A OS=Homo sapiens GN=KRT6A PE=1 SV=3 - [K2C6A_HUMAN] | No | Yes |
| Interferon-induced protein with tetratricopeptide repeats 3 OS=Homo sapiens GN=IFIT3 PE=1 SV=1 - [IFIT3_HUMAN] | No | Yes |
| FAD synthetase OS=Homo sapiens GN=FLAD1 PE=1 SV=1 - [FAD1_HUMAN] | No | Yes |
| Probable alanyl-tRNA synthetase, mitochondrial OS=Homo sapiens GN=AARS2 PE=1 SV=1 - [SYAM_HUMAN] | No | Yes |
| Interferon-induced protein 44-like OS=Homo sapiens GN=IFI44L PE=2 SV=3 - [IF44L_HUMAN] | No | Yes |
| 28S ribosomal protein S5, mitochondrial OS=Homo sapiens GN=MRPS5 PE=1 SV=2 - [RT05_HUMAN] | No | Yes |
| Keratin, type II cytoskeletal 5 OS=Homo sapiens GN=KRT5 PE=1 SV=3 - [K2C5_HUMAN] | No | Yes |
| Acyl-protein thioesterase 1 OS=Homo sapiens GN=LYPLA1 PE=1 SV=1 - [LYPA1_HUMAN] | No | No |
| Receptor-binding cancer antigen expressed on SiSo cells OS=Homo sapiens GN=EBAG9 PE=1 SV=1 - [RCAS1_HUMAN] | No | No |
Western bolt analysis to confirm the results of the proteomic experiment
Through western blotting, we found that serine/threonine-protein kinase1 (AKT1), fermitin family homolog 3 (KIND3, also referred to as URP2), tubulin beta-3 chain (TUBB3) and lactotransferrin (LTF) exhibited higher expression significantly in the cancer group (see Figure 11).
Discussion
Because ovarian cancer patients have few symptoms in the early stages and their tumors become incurable in the late stage, the prognosis of ovarian cancer is worse than that of other common gynecological cancers, such as endometrial cancer and cervical cancer. Therefore, the development of a new method for diagnosing ovarian cancer as early as possible is important for improving the prognosis of this lethal disease. Exosomes are nanoscale vesicles that can be secreted into the extracellular environment, such as blood, urine, and ascites, by both tumor cells and normal cells. Exosomes carry molecular components, such as proteins, mRNAs and miRNAs, derived from their donor cells. Thus, exosomes derived from ovarian cancer patients may carry ovarian cancer-associated bioinformation or characteristics (15). Hence, we hypothesized that exosomes isolated from the peripheral circulation of ovarian cancer patients may have disease-associated molecules that differ from those loaded in exosomes from healthy women, and cancer-derived exosomes may be a potential tool for screening and diagnosing ovarian cancer by analyzing cancer cell-donated components (16,17). Moreover, isolation of exosomes from peripheral circulation requires only a small volume of blood, and this process is not painful for patients, indicating that the use of exosomes as an ovarian cancer diagnostic tool is clinically feasible and patient friendly.
In proteomic studies, either individual samples or pooled samples can be used for analysis. In the present study, after the exosome preparation procedure, we observed some individual samples, mainly from the control group, that did not contain enough protein for further analysis. There are two ways to resolve this problem: gathering a larger volume of blood or using pooled samples. As gathering more blood from study cases may lead to potential harm and ethical conflict, we used pooled samples instead of individual samples for the following proteomic analysis. There are some issues that should be noted when using pooled samples (18). The expression of protein in pooled samples matches the mean expression in individuals who make up the pool, but that may not represent the true expression in individuals. Additionally, the proteins that are detectable in individual samples may not be detected in pooled samples. Diz et al. (18) has proved that using pooled samples in proteomic studies is valid and potentially valuable, but the above issues should be considered in the experimental design.
Because exosomes carry multiple types of proteins, including tumor-associated proteins, several studies (16,19,20) have focused on exploring tumor-associated markers by analyzing exosomal proteins. Traditionally, the analysis of exosomal proteins is performed via 2-dimensional (2D) electrophoresis plus MS. Although this protocol is widely used, it has several shortcomings, such as a poor ability to isolate insoluble proteins and membrane proteins and low detection of proteins showing expression differences of less than two-fold (21). To explore additional exosomal proteins, we used the iTRAQ method plus HPLC/MS in this study. Compared with typical proteomic techniques, iTRAQ uses iron isotopes to label peptides, and the labeled peptides are identified by the subsequent MS/MS procedure directly without electrophoresis. Thus, iTRAQ-based proteomic analysis can identify more proteins/peptides, including extremely acidic or alkaline proteins, which are more difficult to isolate and identify via 2D electrophoresis than by other methods (22). By using iTRAQ plus HPLC/MS, we identified 408 differentially expressed proteins between ovarian cancer exosomes and control exosomes, including 208 downregulated proteins and 200 upregulated proteins in ovarian cancer exosomes; these numbers are much higher than number of exosomal proteins isolated through traditional proteomic analysis (23-25), and the use of high-throughput proteomic analysis is becoming increasingly important in exosome studies (26,27).
Although we identified 200 upregulated proteins in ovarian cancer patient-derived exosomes, we note that these exosomes are not tumor-specific exosomes because the isolation method that we used could not separate exosomes of different origins. We tried to use immunomagnetic beads linked to the tumor-associated marker EpCAM (28,29) and the exosomal marker CD9 (30) to isolate ovarian cancer-specific exosomes. However, it was difficult to obtain enough exosomes for the following proteomic analysis. Furthermore, the exosomal proteins isolated from the peripheral circulation may have multiple origins. To exclude the confounding effects of nontumor cell-derived exosomal proteins, we identified differentially expressed proteins in ovarian cancer tissues and normal epithelial tissues. We believe that the proteins present in both cancer patient-derived exosomes and cancer tissues are cancer-associated proteins and may have tumor biological effects. In this study, 35 proteins were shown to be upregulated in both ovarian cancer patient-derived exosomes and ovarian cancer tissues, which may indicate that these 35 proteins are actual tumor-associated proteins. Specifically, among 35 proteins, there were 15 that were expressed in both ovarian cancer cell lines and exosomes (12) and were detected in the ExoCarta/Vesiclepedia database; thus, these 15 proteins should be further assessed as tumor markers.
Nonexosomal protein contamination should be considered in such analyses. When using the exosome isolation technology applied in our study, high-abundance serum proteins such as albumin, complement proteins and hemoglobin are the main contaminating proteins (31). Because most high-abundance serum proteins do not exist in ovarian tissue, we could exclude serum proteins and confirm whether the exosomal proteins identified in our study came from the ovary by comparing the proteomic results for ovarian tissue. Through this method, we were able to distinguish the ovarian-original proteins from the proteomic results for the exosome, but we could not exclude exosome-unrelated proteins. Thus, we chose the proteins included in the exosomal protein database ExoCarta/Vesiclepedia, and we believe that nonexosome protein contamination could be recognized by using ExoCarta/Vesiclepedia data.
In an analysis of the origin and biological function of these differentially expressed proteins, we found that the upregulated differentially expressed proteins had multiple origins and exhibited various functions in different biological processes. The differentially expressed proteins primarily originated from the cellular membrane and organelles, which might indicate the developmental processes of exosomes (32) and suggest that tumor cells could “throw out” specific components. Interestingly, the top three biological processes for the differentially expressed proteins were “response to stimulus”, “metabolic process” and “immune system process”. These processes may indicate that cancer cells exhibit different responses to stimuli than normal cells through releasing exosomes and that the major biofunctional changes in cancer cells involve the metabolism and interactions of the immune system. However, some circulating exosomes may also come from immune cells, and the different exosomal proteins may indicate that the cancer patients’ immune functions have changed under the influence of cancer cells.
We found that the most common molecular functions of different proteins in circulating exosomes from cancer patients were catalytic activity and binding, which was similar to the findings of our previous study (12). These results indicate that binding proteins on exosomes may help attach exosomes to target cells, and the catalytic activities of the proteins then alter the target cell’s biological behavior. For the protein classes of the different exosomal proteins, nucleic acid-binding proteins and enzyme modulator proteins are the two most common types of proteins following cytoskeletal proteins, which suggests that the different exosomal proteins of cancer patients could bind to target cell nucleic acids and change the enzymatic activities of the target cells. Another possible explanation is that cancer cells may eliminate more nucleic acid-binding or enzyme modulator proteins than normal cells via exosomes to alter their biological activities.
In this study, we found that 35 proteins were upregulated in both ovarian cancer patient serum exosomes and ovarian cancer tissues, indicating that these 35 proteins may be tumor-associated proteins. Among these 35 proteins, 15 proteins were identified in both the ovarian cancer cell lines OVCAR-1/IGROV-1 and the exosome molecular database ExoCarta/Vesiclepedia. Thus, we believe that these 15 proteins should be investigated as potential tumor markers. Hurwitz et al. (14) used 60 cell lines (including 7 ovarian cancer cell lines) to perform a proteomic analysis of EVs. Chloride intracellular channel protein 4, serine/threonine-protein kinase 1, aminoacyl tRNA synthetase complex-interacting multifunctional protein 1, sorting nexin-3, protein FAM49B, fermitin family homolog 3, tubulin beta-3 chain and lactotransferrin were detected in this study, our previous study (12) and by Hurwitz et al. Thioredoxin-related transmembrane protein, thioredoxin domain-containing protein 17, E3 ubiquitin-protein ligase, collagen triple helix repeat-containing protein 1, interferon-induced protein 44-like, 28S mitochondrial ribosomal protein S5, nonspecific lipid-transfer protein, azurocidin, fibrinogen gamma chain, fibrinogen alpha chain, and interferon-induced protein with tetratricopeptide repeats 3 were not detected in our study but were found by Hurwitz et al. Therefore, the 8 proteins detected in both our study and Hurwitz et al.’s study should be further investigated. It should be noted that mass spectrometry is not an accurate method for quantitative protein studies. Therefore, we performed western blotting to confirm whether these eight proteins exhibited higher expression in the cancer group. However, FAM49B, CLIC4, SNX3 and EMAP II were expressed at equal levels in the cancer and control groups. The possible reasons for this result may be that: (I) these proteins are specially modified, such as through phosphorylation, meaning that phosphorylated antibodies would be required for their detection; (II) the peptides detected by mass spectrometry lack sufficient specificity, resulting in inaccurate protein identification; and (III) the concentration of the protein is not suitable. For example, the amount of the t sample may have been either too small or too great for western blotting, meaning that the difference between the two groups could not be accurately judged. In the future, we will use phosphorylated antibodies and different protein concentrations to assess the differences in proteins between the two groups.
Based on reference to THE CANCER GENOME ATLAS (TCGA) and the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) as well as other studies, some of the 8 proteins may be related to ovarian cancer. Serine/threonine-protein kinase 1 is involved in many intracellular signaling pathways. Siu et al. (33) showed that serine/threonine-protein kinase1 is a poor prognostic factor of ovarian cancer. The function of the FAM49b protein is currently unclear, and it may regulate actin dynamics and T cell activation (34) and suppress the proliferation and invasion of cancer cells (35). As a constituent of microtubules, tubulin beta-3 plays a critical role in proper axon guidance and maintenance (36), and its overexpression indicates poor prognosis of ovarian cancer (37). Chloride intracellular channel protein 4 is a trans-membrane ion channel that transports chloride ions and may be related to angiogenesis, promotion of endothelial cell proliferation and regulation of endothelial morphogenesis (38). Based on reference to GEO database GSE18520, which includes 53 advanced-stage, high-grade primary tumor specimens and 10 normal ovarian surface epithelium samples, we found that the genes encoding serine/threonine-protein kinase1 (P<0.01), protein FAM49B (P<0.01), tubulin beta-3 chain (P=0.02) exhibited expression in cancer samples than in normal tissue. Additionally, even though chloride intracellular channel protein 4 expression presented no difference between cancer tissue and normal tissue of GSE 18520, this protein has been proven to serve as a potential ovarian cancer marker (39). Therefore, the four above proteins can be regarded as tumor marker candidates for ovarian cancer for further clinical research. Aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 is noncatalytic component of the multisynthase complex, and its relationship with ovarian cancer has yet to be studied. Sorting nexin-3 is a phosphoinositide-binding protein required for multivesicular body formation (40), and to our knowledge, there are no available reports of the role of this protein in ovarian cancer. By using TCGA data, we found that ovarian cancer patients exhibiting higher gene expression of serine/threonine-protein kinase 1 (HR =1.31, P=0.01), chloride intracellular channel protein 4 (HR =1.19, P=0.01), aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (HR =1.15, P=0.007) or sorting nexin-3 (HR =1.33, P<0.01) presented a worse prognosis. Thus, these four proteins could be further studied as prognostic factors for ovarian cancer. Interestingly, high expression of the tubulin beta-3 chain is a better prognostic factor in TCGA ovarian cancer cases (HR =0.80, P<0.01), but in Ferrandina et al.’s study (37) tubulin beta-3 chain is a poor clinical outcome marker of ovarian cancer, which means that the biological significance of tubulin beta-3 chains for ovarian cancer requires additional study. Fermitin family homolog 3 plays a central role in cell adhesion in hematopoietic cells such as platelets and leukocytes, but there are no available studies on its role in ovarian cancer. Lactotransferrin is usually found in exocrine fluid and functions in antimicrobial activity (41). To the best of our knowledge, the relationship between lactotransferrin and ovarian cancer has yet to be studied. Thus, exploring the relationship of fermitin family homolog 3 and lactotransferrin with ovarian cancer contributes to our knowledge of this fatal tumor.
There are several limitations of this study. (I) In addition to serous carcinoma, which is the most common subtype of EOC, ovarian cancer includes other less common subtypes, such as mucinous carcinoma, clear cell carcinoma, and endometrioid carcinoma. Our study did not include these other subtypes, and the results of this study therefore cannot be extended to all EOC cases. In the future, we could design a study to explore the different exosomal proteins of less common subtypes. (II) The exosomes in the peripheral circulation of EOC patients have multiple origins, which can vary by clinical conditions, and the purification of tumor-specific exosomes from circulating exosomes is difficult using current techniques. In future studies, we could use novel methods such as immunomagnetic bead sorting or tumor-specific antibodies plus flow cytometry to obtain pure tumor-derived exosomes. (III) This work is a retrospective study, and the diagnostic value of the proteins identified in the study for early ovarian cancer is unclear. Therefore, we should design a prospective study to assess the diagnostic value of these eight proteins in the future.
Conclusions
We isolated exosomes from the peripheral blood of EOC patients and noncancer controls and identified 35 proteins that were upregulated in both the EOC patient exosomes and ovarian cancer tissues. Comparisons with exosome molecular databases and other studies identified eight proteins (chloride intracellular channel protein 4, serine/threonine-protein kinase 1, aminoacyl tRNA synthetase complex-interacting multifunctional protein 1, sorting nexin-3, protein FAM49B, fermitin family homolog 3, tubulin beta-3 chain and lactotransferrin) as potential tumor markers, which might offer new tools for the early diagnosis of ovarian cancer.
Acknowledgments
We thank Dr. Hengzi Sun for his assistance in NTA analysis and western blotting.
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Under the rules of the Ethical Committee of our institution, blood sample collection does not require the approval of the committee but must be permitted by the patient. Therefore, before recruitment, all subjects provided signed informed consent and approved their blood samples for use by the researchers.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Buys SS, Partridge E, Black A, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011;305:2295-303. [Crossref] [PubMed]
- Knight AM. Regulated release of B cell-derived exosomes: do differences in exosome release provide insight into different APC function for B cells and DC? Eur J Immunol 2008;38:1186-9. [Crossref] [PubMed]
- Blanc L, De Gassart A, Geminard C, et al. Exosome release by reticulocytes--an integral part of the red blood cell differentiation system. Blood Cells Mol Dis 2005;35:21-6. [Crossref] [PubMed]
- Zhao Z, Yang Y, Zeng Y, et al. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016;16:489-96. [Crossref] [PubMed]
- Schey KL, Luther JM, Rose KL. Proteomics characterization of exosome cargo. Methods 2015;87:75-82. [Crossref] [PubMed]
- Yentrapalli R, Merl-Pham J, Azimzadeh O, et al. Quantitative changes in the protein and miRNA cargo of plasma exosome-like vesicles after exposure to ionizing radiation. Int J Radiat Biol 2017;93:569-80. [Crossref] [PubMed]
- Yang J, Wei F, Schafer C, et al. Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS One 2014;9:e110641. [Crossref] [PubMed]
- Kelleher RJ Jr, Balu-Iyer S, Loyall J, et al. Extracellular Vesicles Present in Human Ovarian Tumor Microenvironments Induce a Phosphatidylserine-Dependent Arrest in the T-cell Signaling Cascade. Cancer Immunol Res 2015;3:1269-78. [Crossref] [PubMed]
- Meng X, Muller V, Milde-Langosch K, et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016;7:16923-35. [Crossref] [PubMed]
- Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol Rep 2011;25:749-62. [PubMed]
- Liang B, Peng P, Chen S, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics 2013;80:171-82. [Crossref] [PubMed]
- Zhang W, Peng P, Kuang Y, et al. Characterization of exosomes derived from ovarian cancer cells and normal ovarian epithelial cells by nanoparticle tracking analysis. Tumour Biol 2016;37:4213-21. [Crossref] [PubMed]
- Hurwitz SN, Rider MA, Bundy JL, et al. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016;7:86999-7015. [Crossref] [PubMed]
- Kobayashi M, Salomon C, Tapia J, et al. Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200. J Transl Med 2014;12:4. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Fais S, O'Driscoll L, Borras FE, et al. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano 2016;10:3886-99. [Crossref] [PubMed]
- Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis 2009;30:2967-75. [Crossref] [PubMed]
- Cappello F, Logozzi M, Campanella C, et al. Exosome levels in human body fluids: A tumor marker by themselves? Eur J Pharm Sci 2017;96:93-8. [Crossref] [PubMed]
- Zocco D, Ferruzzi P, Cappello F, et al. Extracellular vesicles as shuttles of tumor biomarkers and anti-tumor drugs. Front Oncol 2014;4:267. [Crossref] [PubMed]
- Klein E, Klein JB, Thongboonkerd V. Two-dimensional gel electrophoresis: a fundamental tool for expression proteomics studies. Contrib Nephrol 2004;141:25-39. [Crossref] [PubMed]
- Evans C, Noirel J, Ow SY, et al. An insight into iTRAQ: where do we stand now? Anal Bioanal Chem 2012;404:1011-27. [Crossref] [PubMed]
- Théry C, Boussac M, Veron P, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol 2001;166:7309-18. [Crossref] [PubMed]
- Wubbolts R, Leckie RS, Veenhuizen PT, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem 2003;278:10963-72. [Crossref] [PubMed]
- Mears R, Craven RA, Hanrahan S, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 2004;4:4019-31. [Crossref] [PubMed]
- Sinha A, Ignatchenko V, Ignatchenko A, et al. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem Biophys Res Commun 2014;445:694-701. [Crossref] [PubMed]
- Jeppesen DK, Nawrocki A, Jensen SG, et al. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics 2014;14:699-712. [Crossref] [PubMed]
- Vaksman O, Trope C, Davidson B, et al. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 2014;35:2113-20. [Crossref] [PubMed]
- Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 2015;136:2616-27. [Crossref] [PubMed]
- Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012;56:293-304. [Crossref] [PubMed]
- Diaz G, Bridges C, Lucas M, et al. Protein Digestion, Ultrafiltration, and Size Exclusion Chromatography to Optimize the Isolation of Exosomes from Human Blood Plasma and Serum. J Vis Exp 2018; [Crossref] [PubMed]
- Bobrie A, Colombo M, Raposo G, et al. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 2011;12:1659-68. [Crossref] [PubMed]
- Siu MK, Wong ES, Chan HY, et al. Differential expression and phosphorylation of Pak1 and Pak2 in ovarian cancer: effects on prognosis and cell invasion. Int J Cancer 2010;127:21-31. [Crossref] [PubMed]
- Shang W, Jiang Y, Boettcher M, et al. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc Natl Acad Sci U S A 2018;115:E4051-60. [Crossref] [PubMed]
- Chattaragada MS, Riganti C, Sassoe M, et al. FAM49B, a novel regulator of mitochondrial function and integrity that suppresses tumor metastasis. Oncogene 2018;37:697-709. [Crossref] [PubMed]
- Tischfield MA, Baris HN, Wu C, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 2010;140:74-87. [Crossref] [PubMed]
- Ferrandina G, Zannoni GF, Martinelli E, et al. Class III beta-tubulin overexpression is a marker of poor clinical outcome in advanced ovarian cancer patients. Clin Cancer Res 2006;12:2774-9. [Crossref] [PubMed]
- Tung JJ, Hobert O, Berryman M, et al. Chloride intracellular channel 4 is involved in endothelial proliferation and morphogenesis in vitro. Angiogenesis 2009;12:209-20. [Crossref] [PubMed]
- Tang HY, Beer LA, Chang-Wong T, et al. A xenograft mouse model coupled with in-depth plasma proteome analysis facilitates identification of novel serum biomarkers for human ovarian cancer. J Proteome Res 2012;11:678-91. [Crossref] [PubMed]
- Pons V, Luyet PP, Morel E, et al. Hrs and SNX3 functions in sorting and membrane invagination within multivesicular bodies. PLoS Biol 2008;6:e214. [Crossref] [PubMed]
- Sánchez L, Calvo M, Brock JH. Biological role of lactoferrin. Arch Dis Child 1992;67:657-61. [Crossref] [PubMed]