
miR-1290 inhibits chordoma cell proliferation and invasion by targeting Robo1
Introduction
Chordoma is a rare and malignant bone tumor that arises from the remnants of the embryonic notochord (1). It mostly occurs at the sacrum, skull base and mobile spine (1). Chordoma is typically a low grade but remains a locally invasive malignant neoplasm (2,3). Currently, surgery remains the most effective treatment as the tumor is largely resistant to chemotherapy and radiotherapy (4). However, it still has high recurrence and metastasis after surgical resection (5). Therefore, the study of the mechanisms for chordoma development is an urgent necessity.
MicroRNAs (miRNAs), which are small non-coding single-stranded RNA molecules of about 18–25 nucleotides long, can regulate gene expression and function by mainly interacting with the 3'-UTR of their target mRNA (6). Increasing evidence has shown that miRNA dysregulation plays a critical role in tumor initiation and progression. They can act as tumor suppressors or oncogenes in numerous cancer types, including chordoma (7-9). For example, miR-1 was reported to be downregulated in chordoma and showed its inhibition effect in cell migration and invasion by targeting the gene slug in chordoma (10). Meanwhile, reduced miR-1237-3p expression was shown to be related with chordoma invasion and a worse recurrence-free survival of patients (11).
Recently, some reports have shown that miR-1290 is significantly involved in tumor invasion and proliferation (12-14). Our previous studies, by using miRNA array, demonstrated that miR-1290 was significantly down-regulated in chordoma compared to fetal notochord pulposus (FNP) (15). Survival analysis indicated that low expression of miR-1290 was significantly correlated with the invasion and recurrence of chordoma (16). However, the underlying mechanisms by which miR-1290 functions via target genes in chordoma remain unclear. Thus, further identification and validation of novel targets of miR-1290 is important for understanding the development of chordoma. In this study, we found that the over-expression of miR-1290 inhibited chordoma cell invasion and proliferation in vitro through Robo1, which is a miR-1290 target gene.
Methods
Tissue specimens
Sixteen sacral chordoma samples were collected from January 2010 to December 2016 at the First Affiliated Hospital of Soochow University (Suzhou, China). Ten fetal nucleus pulposus specimens were obtained from aborted fetuses with a gestational age of 8–24 weeks in the Department of Gynecology and Obstetrics. Parts of the samples were fixed in a 10% formalin solution and embedded in paraffin. Histologic sections were stained with hematoxylin and eosin for histology diagnosis. The other parts of the specimens were collected and preserved in liquid nitrogen. All the patients or their families were informed, and supplied standard written consent. Our study protocol was approved by the Ethics Review Committee of the Institutional Review Board of the hospital.
Immunohistochemistry (IHC)
Immunohistochemical staining was carried out with an EnVision two-step staining method on 4-µm-thick tissue sections (17). The sections were dewaxed in xylene and rehydrated in ethanol before the antigen retrieval. The primary antibody (anti-Robo1, dilution at 1/50, Abcam) and secondary antibody were used. For positive control, tissue sections of human hepatocarcinoma with known positivity were used in each batch of staining. For negative control (NC), sections with phosphate-buffered saline were treated with an omission of the primary antibody. Immunostaining was evaluated by two independent pathologists blinded to clinical information. Tissue specimens were scored according to the color intensity and the number of positive cells. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown) or 3 (brown), and the number of positive cells was graded as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (>50%). The two grades were added together, and specimens were assigned to low expression [0–2], or high expression [3–6].
Cell culture and transfection
Human chordoma cell line U-CH1 was obtained from American type culture collection (ATCC). Cells were cultured in IMDM, and RPMI1640 (1:4) supplemented with 10% fetal bovine serum and penicillin/streptomycin (100 U/mL) (Invitrogen, CA, USA). Cells were cultured in an incubator containing 5% CO2 at 37 °C. The medium was changed every 3 days and cells were subcultured once in confluence. miR-1290 mimic, miR-1290 mimic NC, miR-1290 inhibitor, miR-1290 inhibitor NC, and siRobo1 were each transfected into chordoma cells using Lipofectamine 2000 reagent (Invitrogen, CA, USA) according to the manufacturer’s instruction. After 48 h transfection, the cells were harvested for subsequent experimental studies.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
For miR-1290 detection, miRNA was extracted from clinical samples and U-CH1 cells using RNA extraction kit (Applied Biosystems; Life Technologies). The cDNA was synthesized using the TaqMan miRNA reverse transcription kit (Applied Biosystems; Life Technologies) following the manufacturer’s instructions. qRT-PCR was performed using TaqMan microRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.). Primer sequences used in this study were as follows: miR-1290, forward 5′-CAGTGCTGGATTTTTGGAT-3′, reverse 5′-TATGGTTGTTCACGACTCCTTCAC-3′; U6, forward 5′-ATTGGAACGATACAGAGAAGATT-3′, reverse 5′-GGAACGCTTCACGAATTTG-3′. Robo1 siRNA, forward 5′-GGAUGUAUUUGCAACAAGATT-3′, reverse 5′-GGAUGUAUUUGCA ACAA GATT-3′; scramble siRNA, forward 5′-UUCUCCGAACGUGUCACGUTT-3′, reverse 5′-ACGUGACACGUUCGGAGAATT-3′.
Western blot analysis
The whole cell lysate from the U-CH1 cell line was extracted using a radioimmunoprecipitation assay (RIPA) buffer. Proteins were separated on 10% sodium dodecyl sulfate (SDS) polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking with 5% non-fat milk in tris-buffered saline-tween (TBST) for 1 h, the membranes were immersed with the primary antibody (anti-Robo1, 1/50 dilution, Abcam) at 4 °C overnight. Membranes were then washed and incubated with secondary antibodies at room temperature. GAPDH (1:1,000, Cell Signaling) was used as an internal loading control.
Cell clone formation assay
Cells lines transfected with the miR-1290 mimic, inhibitor, siRobo1 or control were plated in duplicate in a 6-well plate pre-treated with type I rat tail collagen (BD biotechnology). After incubation at 37 °C for 14 days, the colonies were stained with crystal Violet solution in methanol for 15 min.
Transwell assay
After 48 h transfection, Transwell assay was used to evaluate cellular invasion. 2×105 U-CH1 cells were suspended in serum-free DMEM and then seeded into the upper chamber of each insert coated with Matrigel, while the lower chamber was filled with DMEM supplemented with 20% FBS. After 48 h of incubation, cells on the bottom side of the membrane were fixed with 4% polyoxymethylene for 15 minutes and stained with crystal violet dye for 5–10 minutes at 37 °C and finally visualized by using a light microscope (Carl Zeiss, Germany).
Luciferase reporter assay
The Robo1 3′-UTR containing miR-1290 binding sequences and its MUT 3′-UTR were cloned into pGL3-promoter vector (Promega, Madison, WI, USA). For luciferase reporter assay, U-CH1 cells were co-transfected with pGL3-Robo1 3′-UTR-MUT and miR1290 mimic or NC using Lipofectamine 2000. Forty-eight hours after transfection, luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega, USA). Renilla luciferase activity was used to normalize for transfection efficiency.
Statistical analyses
SPSS 16.0 software (Chicago, IL, USA) was used for statistical analyses. Numerical data were expressed as mean ± standard deviation. Comparisons of quantitative data between the two groups were performed using two-sided Student’s t-test, and the rates were compared using a Chi-square test. The statistical association between miR-1290 and Robo1 in chordoma tissue was evaluated with Spearman’s rank correlation test. A P value <0.05 was considered as significant, and a P value <0.01 was considered as highly significant.
Results
The expression of miR-1290 and Robo1 in chordoma tissues
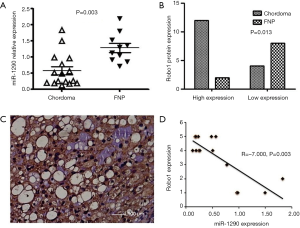
miR-1290 expression levels in chordoma tissue and FNP were detected by qRT-PCR. miR-1290 expression was significantly down-regulated in chordoma tissue compared to FNP tissue (Figure 1A). In the meantime, we performed IHC in the chordoma tissues to identify the expression of Robo1. The expression of Robo1 in the chordoma was significantly higher than that in the FNP (75% vs. 20%, P=0.013) (Figure 1B,C). Correlation analysis showed that there was a negative correlation between the expression of miR-1290 and Robo1 in chordoma tissue (Figure 1D).
miR-1290 suppresses chordoma cells proliferation and invasion in vitro
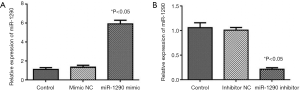
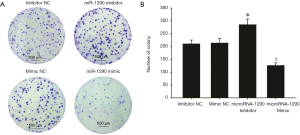
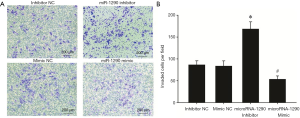
U-CH1 cells were transfected with miR-1290 mimics or miR-1290 inhibitor to explore the role of miR-1290 in chordoma progression. qRT-PCR analysis showed that miR-1290 was upregulated in the miR-1290 mimic transfection group and downregulated in the miR-1290 inhibitor transfection group (P<0.05) (Figure 2). Cell clone formation assay showed that the number of cell clones in the miR-1290 mimic group was significantly lower than that in the miR-1290 mimic NC group (P<0.05), while the number in the miR-1290 inhibitor group was significantly higher than that in the miR-1290 inhibitor NC group (P<0.05) (Figure 3). Transwell invasion test showed that the number of cells that passed through the membrane in the miR-1290 inhibitor group was significantly higher than that in the miR-1290 inhibitor NC group (P<0.05), while the number of cells in the miR-1290 mimic group was significantly lowered than that in the miR-1290 mimic NC group (P<0.05) (Figure 4).
Validation of Robo1 as a direct target of miR-1290
Bioinformatics analysis was performed to understand further the mechanisms underlying the role of miR-1290 in chordoma. The target genes of miRNA-1290 were predicted by using the TargetScan, microRNAorg and PITA databases. Robo1 was identified as a likely target for miR-1290 because there was a complementarity relationship between miR-1290 and the Robo1 3′-UTR (Figure 5A). The results of the Western blot analysis showed that the expression of Robo1 was significantly increased in the miR-1290 inhibitor transfection group, while it decreased significantly in the miR-1290 mimic transfection group (Figure 5B,C). The dual luciferase reporter assay showed that upregulation of miR-1290 significantly reduced the firefly luciferase activity of Robo1-3′-UTR (P<0.05) (Figure 5D,E).
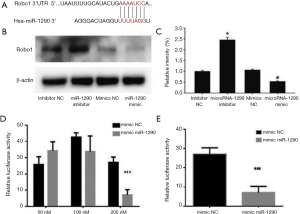
miR-1290 inhibits chordoma cell proliferation and invasion by suppressing Robo1 expression
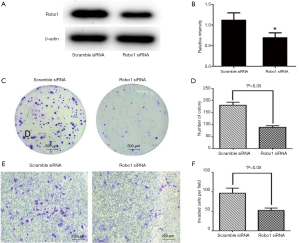
U-CH1 cell line was transfected with siRobo1 and miR-1290 inhibitors or miR-1290 mimics to explore the underlying mechanism of miR-1290 and Robo1 in chordoma cell proliferation and invasion. Western blot results showed that the transfection of siRobo1 could significantly reduce the expression of Robo1 protein in chordoma cells. The cell cloning formation assay and Transwell invasion experiment results showed that downregulation of Robo1 significantly inhibited the proliferation and invasion ability of chordoma cells (Figure 6). The results of cell cloning formation assay showed that the proliferation ability of the Robo1 siRNA + miR-1290 inhibitor group was significantly higher than that of the Robo1 siRNA + inhibitor NC group (P<0.05), while the proliferation ability was significantly lower in the Robo1 siRNA + miR-1290 mimic group than that of the Robo1 siRNA + mimic NC co-transfection group (P<0.05) (Figure 7A,B). Transwell invasion assay results showed that the invasion ability of the Robo1 siRNA + miR-1290 inhibitor group was significantly higher than that of the Robo1 siRNA + inhibitor NC co-transfection group (P<0.05), while the invasion ability of the Robo1 siRNA + miR-1290 mimic group was significantly lower than that of the Robo1 siRNA + mimic NC co-transfection group (P<0.05) (Figure 7C,D).

Discussion
Chordomas are the fourth most common malignant neoplasms originating from bone (4). Although histologically low-grade, they are highly invasive and often recur locally (4). The gold standard for the treatment of chordoma is a radical en bloc resection with wide margins (1,4). Nevertheless, large en bloc resection is feasible for less than 50% of sacral chordomas and even fewer clival chordomas (18). Moreover, large en bloc resection may require extensive nerve and ligamentous excision leading to ambulatory, sexual, bowel and urinary incontinence (19). The overall 5- and 10-year survival rates following sacrectomy are 45–77% and 28–50% respectively (4).
Exploring detailed molecular mechanisms in chordoma progression and providing novel therapeutic targets are urgently needed. Recently, miR-1290 was reported to have a great role in tumor progression (12,20,21). Endo et al. reported that miR-1290 was down-regulated in ER high Ki67 low breast cancer and might be associated with its characteristics (12). Li et al. revealed that miR-1290 was elevated and may be correlated with cell proliferation and invasion in esophageal squamous cell carcinoma (14). Cancer stem cells-associated miR-1246 or miR-1290 are important in the invasion or metastasis of non-small cell lung cancer (22). Therefore, we can see that miR-1290 plays various roles in different kinds of cancers.
Our previous studies have shown that miR-1290 was down-regulated in chordoma tissues and might be negatively related to the prognosis of chordoma (15,16). However, the real underlying mechanisms are unclear. Therefore, we explored the specific functions of miR-1290 in chordoma in this study. After transfected with miR-1290 mimics, chordoma cell proliferation and invasion were both significantly inhibited, while the opposite effects were observed when transfected with miR-1290 inhibitor. These all indicate that miR-1290 might act as a tumor suppressor in chordoma.
To probe the mechanism by which miR-1290 influences chordoma cells, bioinformatics tools were used in our study. Robo1 was identified as a potential target gene of miR-1290. Accumulating evidence has shown that Robo1 can promote tumor angiogenesis and migration in many types of cancers (23-25). In our study, Robo1 expression was found to be up-regulated in chordoma by using IHC. We also observed that expression of Robo1 was negatively correlated with miR-1290 in chordoma tissue. Moreover, Robo1 expression was significantly increased when miR-1290 inhibitor was transfected into the U-CH1 cell line. Furthermore, by using luciferase reporter assay, we confirmed that Robo1 is a direct target of miR-1290 in chordoma cells.
Robo1 has been reported to be over-expressed in many cancers, and its dysregulation has great importance in tumor invasion and angiogenesis (24-26). Zhou et al. found that Robo1 and Slit2 were both overexpressed in colorectal epithelial carcinoma, and that knockdown of endogenous Robo1 could diminish tumor growth and liver metastasis (27). miR-218 elevation was discovered to inhibit cell invasion and migration by downregulating Robo1 in gastric cancer (28). Dontula et al. reported that miR-203 could inhibit migration of the glioma cells by disrupting the Robo1/ERK/MMP-9 signaling axis (29). Consistently, in the current study we found that downregulating miR-1290 could restore the inhibitory effect of Robo1 downregulation in chordoma cells. Meanwhile, elevating miR-1290 could facilitate the effect of Robo1 downregulation in chordoma cells.
In conclusion, we discovered that miR-1290 is a tumor suppressor in chordoma, and it can inhibit the chordoma cell’s proliferation and invasion by negatively regulating the direct target gene Robo1. Although the exact molecular mechanism remains to be further explored, our present study is the first to report how miR-1290 might inhibit chordoma progression by targeting Robo1, which provides new clues for guiding therapeutic intervention in chordoma.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All the patients or their families were informed, and supplied standard written consent. Our study protocol was approved by the Ethics Review Committee of the Institutional Review Board of the hospital (Approval ID: 185-1).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Casali PG, Stacchiotti S, Sangalli C, et al. Chordoma. Curr Opin Oncol 2007;19:367-70. [Crossref] [PubMed]
- Bergh P, Kindblom LG, Gunterberg B, et al. Prognostic factors in chordoma of the sacrum and mobile spine: a study of 39 patients. Cancer 2000;88:2122-34. [Crossref] [PubMed]
- Schwab JH, Boland PJ, Agaram NP, et al. Chordoma and chondrosarcoma gene profile: implications for immunotherapy. Cancer Immunol Immunother 2009;58:339-49. [Crossref] [PubMed]
- Kayani B, Hanna SA, Sewell MD, et al. A review of the surgical management of sacral chordoma. Eur J Surg Oncol 2014;40:1412-20. [Crossref] [PubMed]
- Walcott BP, Nahed BV, Mohyeldin A, et al. Chordoma: current concepts, management, and future directions. Lancet Oncol 2012;13:e69-76. [Crossref] [PubMed]
- Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle 2008;7:2485-92. [Crossref] [PubMed]
- Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009;58:1375-81. [Crossref] [PubMed]
- Duan Z, Choy E, Nielsen GP, et al. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res 2010;28:746-52. [PubMed]
- Kong YW, Ferland-McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol 2012;13:e249-58. [Crossref] [PubMed]
- Osaka E, Yang X, Shen JK, et al. MicroRNA-1 (miR-1) inhibits chordoma cell migration and invasion by targeting slug. J Orthop Res 2014;32:1075-82. [Crossref] [PubMed]
- Zou MX, Huang W, Wang XB, et al. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J 2015;24:1738-46. [Crossref] [PubMed]
- Endo Y, Toyama T, Takahashi S, et al. miR-1290 and its potential targets are associated with characteristics of estrogen receptor alpha-positive breast cancer. Endocr Relat Cancer 2013;20:91-102. [Crossref] [PubMed]
- Mo D, Gu B, Gong X, et al. miR-1290 is a potential prognostic biomarker in non-small cell lung cancer. J Thorac Dis 2015;7:1570-9. [PubMed]
- Li M, He XY, Zhang ZM, et al. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol 2015;21:3245-55. [Crossref] [PubMed]
- Chen K, Chen H, Zhang K, et al. MicroRNA profiling and bioinformatics analyses reveal the potential roles of microRNAs in chordoma. Oncol Lett 2017;14:5533-9. [PubMed]
- Zhang K, Chen H, Zhang B, et al. Overexpression of Raf-1 and ERK1/2 in sacral chordoma and association with tumor recurrence. Int J Clin Exp Pathol 2015;8:608-14. [PubMed]
- Sabattini E, Bisgaard K, Ascani S, et al. The EnVision++ system: a new immunohistochemical method for diagnostics and research. Critical comparison with the APAAP, ChemMate, CSA, LABC, and SABC techniques. J Clin Pathol 1998;51:506-11. [Crossref] [PubMed]
- Lebellec L, Aubert S, Zairi F, et al. Molecular targeted therapies in advanced or metastatic chordoma patients: facts and hypotheses. Crit Rev Oncol Hematol 2015;95:125-31. [Crossref] [PubMed]
- Hulen CA, Temple HT, Fox WP, et al. Oncologic and functional outcome following sacrectomy for sacral chordoma. J Bone Joint Surg Am 2006;88:1532-9. [Crossref] [PubMed]
- Janiszewska J, Szaumkessel M, Kostrzewska-Poczekaj M, et al. Global miRNA Expression Profiling Identifies miR-1290 as Novel Potential oncomiR in Laryngeal Carcinoma. PLoS One 2015;10:e0144924. [Crossref] [PubMed]
- Mao Y, Liu J, Zhang D, et al. MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother 2015;76:82-93. [Crossref] [PubMed]
- Zhang WC, Chin TM, Yang H, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun 2016;7:11702. [Crossref] [PubMed]
- Ao JY, Chai ZT, Zhang YY, et al. Robo1 promotes angiogenesis in hepatocellular carcinoma through the Rho family of guanosine triphosphatases' signaling pathway. Tumour Biol 2015;36:8413-24. [Crossref] [PubMed]
- Villacis RA, Abreu FB, Miranda PM, et al. ROBO1 deletion as a novel germline alteration in breast and colorectal cancer patients. Tumour Biol 2016;37:3145-53. [Crossref] [PubMed]
- He H, Hao SJ, Yao L, et al. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol Ther 2014;15:1333-9. [Crossref] [PubMed]
- Jiang L, Wang Y, Rong Y, et al. miR-1179 promotes cell invasion through SLIT2/ROBO1 axis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol 2015;8:319-27. [PubMed]
- Zhou WJ, Geng ZH, Chi S, et al. Slit-Robo signaling induces malignant transformation through Hakai-mediated E-cadherin degradation during colorectal epithelial cell carcinogenesis. Cell Res 2011;21:609-26. [Crossref] [PubMed]
- Tie J, Pan Y, Zhao L, et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet 2010;6:e1000879. [Crossref] [PubMed]
- Dontula R, Dinasarapu A, Chetty C, et al. MicroRNA 203 Modulates Glioma Cell Migration via Robo1/ERK/MMP-9 Signaling. Genes Cancer 2013;4:285-96. [Crossref] [PubMed]


